On origin of genetic code and tRNA before translation
- PMID: 21342520
- PMCID: PMC3050877
- DOI: 10.1186/1745-6150-6-14
On origin of genetic code and tRNA before translation
Abstract
Background: Synthesis of proteins is based on the genetic code - a nearly universal assignment of codons to amino acids (aas). A major challenge to the understanding of the origins of this assignment is the archetypal "key-lock vs. frozen accident" dilemma. Here we re-examine this dilemma in light of 1) the fundamental veto on "foresight evolution", 2) modular structures of tRNAs and aminoacyl-tRNA synthetases, and 3) the updated library of aa-binding sites in RNA aptamers successfully selected in vitro for eight amino acids.
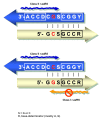
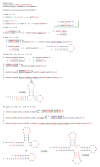
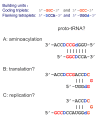
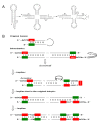
Results: The aa-binding sites of arginine, isoleucine and tyrosine contain both their cognate triplets, anticodons and codons. We have noticed that these cases might be associated with palindrome-dinucleotides. For example, one-base shift to the left brings arginine codons CGN, with CG at 1-2 positions, to the respective anticodons NCG, with CG at 2-3 positions. Formally, the concomitant presence of codons and anticodons is also expected in the reverse situation, with codons containing palindrome-dinucleotides at their 2-3 positions, and anticodons exhibiting them at 1-2 positions. A closer analysis reveals that, surprisingly, RNA binding sites for Arg, Ile and Tyr "prefer" (exactly as in the actual genetic code) the anticodon(2-3)/codon(1-2) tetramers to their anticodon(1-2)/codon(2-3) counterparts, despite the seemingly perfect symmetry of the latter. However, since in vitro selection of aa-specific RNA aptamers apparently had nothing to do with translation, this striking preference provides a new strong support to the notion of the genetic code emerging before translation, in response to catalytic (and possibly other) needs of ancient RNA life. Consistently with the pre-translation origin of the code, we propose here a new model of tRNA origin by the gradual, Fibonacci process-like, elongation of a tRNA molecule from a primordial coding triplet and 5'DCCA3' quadruplet (D is a base-determinator) to the eventual 76 base-long cloverleaf-shaped molecule.
Conclusion: Taken together, our findings necessarily imply that primordial tRNAs, tRNA aminoacylating ribozymes, and (later) the translation machinery in general have been co-evolving to ''fit'' the (likely already defined) genetic code, rather than the opposite way around. Coding triplets in this primal pre-translational code were likely similar to the anticodons, with second and third nucleotides being more important than the less specific first one. Later, when the code was expanding in co-evolution with the translation apparatus, the importance of 2-3 nucleotides of coding triplets "transferred" to the 1-2 nucleotides of their complements, thus distinguishing anticodons from codons. This evolutionary primacy of anticodons in genetic coding makes the hypothesis of primal stereo-chemical affinity between amino acids and cognate triplets, the hypothesis of coding coenzyme handles for amino acids, the hypothesis of tRNA-like genomic 3' tags suggesting that tRNAs originated in replication, and the hypothesis of ancient ribozymes-mediated operational code of tRNA aminoacylation not mutually contradicting but rather co-existing in harmony.
Figures





Similar articles
-
The Uroboros Theory of Life's Origin: 22-Nucleotide Theoretical Minimal RNA Rings Reflect Evolution of Genetic Code and tRNA-rRNA Translation Machineries.Acta Biotheor. 2019 Dec;67(4):273-297. doi: 10.1007/s10441-019-09356-w. Epub 2019 Aug 6. Acta Biotheor. 2019. PMID: 31388859
-
One ancestor for two codes viewed from the perspective of two complementary modes of tRNA aminoacylation.Biol Direct. 2009 Jan 27;4:4. doi: 10.1186/1745-6150-4-4. Biol Direct. 2009. PMID: 19173731 Free PMC article.
-
Deamination gradients within codons after 1<->2 position swap predict amino acid hydrophobicity and parallel β-sheet conformational preference.Biosystems. 2020 May;191-192:104116. doi: 10.1016/j.biosystems.2020.104116. Epub 2020 Feb 18. Biosystems. 2020. PMID: 32081715
-
Origin and Evolution of the Universal Genetic Code.Annu Rev Genet. 2017 Nov 27;51:45-62. doi: 10.1146/annurev-genet-120116-024713. Epub 2017 Aug 30. Annu Rev Genet. 2017. PMID: 28853922 Review.
-
The origin of the genetic code: amino acids as cofactors in an RNA world.Trends Genet. 1999 Jun;15(6):223-9. doi: 10.1016/s0168-9525(99)01730-8. Trends Genet. 1999. PMID: 10354582 Review.
Cited by
-
A Model for the Origin of the First mRNAs.J Mol Evol. 2015 Aug;81(1-2):10-7. doi: 10.1007/s00239-015-9691-y. Epub 2015 Jul 24. J Mol Evol. 2015. PMID: 26205159
-
Amino acid fermentation at the origin of the genetic code.Biol Direct. 2012 Feb 10;7:6. doi: 10.1186/1745-6150-7-6. Biol Direct. 2012. PMID: 22325238 Free PMC article.
-
Transfer RNAs: diversity in form and function.RNA Biol. 2021 Mar;18(3):316-339. doi: 10.1080/15476286.2020.1809197. Epub 2020 Sep 9. RNA Biol. 2021. PMID: 32900285 Free PMC article. Review.
-
The Origin of Translation: Bridging the Nucleotides and Peptides.Int J Mol Sci. 2022 Dec 22;24(1):197. doi: 10.3390/ijms24010197. Int J Mol Sci. 2022. PMID: 36613641 Free PMC article. Review.
-
The Rodin-Ohno hypothesis that two enzyme superfamilies descended from one ancestral gene: an unlikely scenario for the origins of translation that will not be dismissed.Biol Direct. 2014 Jun 14;9:11. doi: 10.1186/1745-6150-9-11. Biol Direct. 2014. PMID: 24927791 Free PMC article.
References
-
- Maynard Smith J, Szathmáry E. W.H. Freeman spectrum. Oxford, New York, Heidelberg; 1995. The major transitions in evolution.
-
- Woese CR. In: Ribosomes: Structure, Function, and Genetics. Chambliss G, et al, editor. University Park Press; 1980. Just So Stories and Rube Goldberg machines: speculations on the origin of the protein synthetic machinery; pp. 357–373.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

