Regulation of gene expression by reiterative transcription
- PMID: 21334966
- PMCID: PMC3078970
- DOI: 10.1016/j.mib.2011.01.012
Regulation of gene expression by reiterative transcription
Abstract
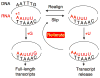
Gene regulation involves many different types of transcription control mechanisms, including mechanisms based on reiterative transcription in which nucleotides are repetitively added to the 3' end of a nascent transcript due to upstream transcript slippage. In these mechanisms, reiterative transcription is typically modulated by interactions between RNA polymerase and its nucleoside triphosphate substrates without the involvement of regulatory proteins. This review describes the current state of knowledge of gene regulation involving reiterative transcription. It focuses on the methods by which reiterative transcription is controlled and emphasizes the different fates of transcripts produced by this reaction. The review also includes a discussion of possible new and fundamentally different mechanisms of gene regulation that rely on conditional reiterative transcription.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
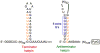

Similar articles
-
Transcription start site sequence and spacing between the -10 region and the start site affect reiterative transcription-mediated regulation of gene expression in Escherichia coli.J Bacteriol. 2014 Aug 15;196(16):2912-20. doi: 10.1128/JB.01753-14. Epub 2014 Jun 2. J Bacteriol. 2014. PMID: 24891446 Free PMC article.
-
X-ray crystal structure of a reiterative transcription complex reveals an atypical RNA extension pathway.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8211-8216. doi: 10.1073/pnas.1702741114. Epub 2017 Jun 26. Proc Natl Acad Sci U S A. 2017. PMID: 28652344 Free PMC article.
-
Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching.J Mol Biol. 1995 Dec 8;254(4):552-65. doi: 10.1006/jmbi.1995.0638. J Mol Biol. 1995. PMID: 7500333
-
Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors.Microbiol Mol Biol Rev. 2008 Jun;72(2):266-300, table of contents. doi: 10.1128/MMBR.00001-08. Microbiol Mol Biol Rev. 2008. PMID: 18535147 Free PMC article. Review.
-
In vitro assay for reiterative transcription during transcriptional initiation by Escherichia coli RNA polymerase.Methods Enzymol. 1996;273:71-85. doi: 10.1016/s0076-6879(96)73007-0. Methods Enzymol. 1996. PMID: 8791600 Review. No abstract available.
Cited by
-
The fidelity of transcription: RPB1 (RPO21) mutations that increase transcriptional slippage in S. cerevisiae.J Biol Chem. 2013 Jan 25;288(4):2689-99. doi: 10.1074/jbc.M112.429506. Epub 2012 Dec 5. J Biol Chem. 2013. PMID: 23223234 Free PMC article.
-
Initiating nucleotide identity determines efficiency of RNA synthesis from 6S RNA templates in Bacillus subtilis but not Escherichia coli.Nucleic Acids Res. 2013 Aug;41(15):7501-11. doi: 10.1093/nar/gkt517. Epub 2013 Jun 12. Nucleic Acids Res. 2013. PMID: 23761441 Free PMC article.
-
Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology.FEMS Microbiol Rev. 2019 May 1;43(3):304-339. doi: 10.1093/femsre/fuz001. FEMS Microbiol Rev. 2019. PMID: 30721976 Free PMC article. Review.
-
Differential role of base pairs on gal promoters strength.J Mol Biol. 2015 Feb 27;427(4):792-806. doi: 10.1016/j.jmb.2014.12.010. Epub 2014 Dec 24. J Mol Biol. 2015. PMID: 25543084 Free PMC article.
-
Regulation of Bacterial Gene Expression by Transcription Attenuation.Microbiol Mol Biol Rev. 2019 Jul 3;83(3):e00019-19. doi: 10.1128/MMBR.00019-19. Print 2019 Aug 21. Microbiol Mol Biol Rev. 2019. PMID: 31270135 Free PMC article. Review.
References
-
- Jacques JP, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991;5:707–713. - PubMed
-
- Linton MF, Raabe M, Pierotti V, Young SG. Reading-frame restoration by transcriptional slippage at long stretches of adenine residues in mammalian cells. J Biol Chem. 1997;272:14127. - PubMed
-
- Martin CT, Muller DK, Coleman JE. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988;27:3966–3974. - PubMed
-
- Qi F, Turnbough CL., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. This paper is the original report of codBA regulation by reiterative transcription in E. coli, which is used in this review as the example of gene regulation involving indirect control of reiterative transcription. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

