Selective activation of the transcription factor NFAT1 by calcium microdomains near Ca2+ release-activated Ca2+ (CRAC) channels
- PMID: 21325277
- PMCID: PMC3083208
- DOI: 10.1074/jbc.M111.220582
Selective activation of the transcription factor NFAT1 by calcium microdomains near Ca2+ release-activated Ca2+ (CRAC) channels
Abstract
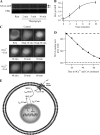
NFATs are a family of Ca(2+)-dependent transcription factors that play a central role in the morphogenesis, development, and physiological activities of numerous distinct cell types and organ systems. Here, we visualize NFAT1 movement in and out of the nucleus in response to transient activation of store-operated Ca(2+) release-activated Ca(2+) (CRAC) channels in nonexcitable cells. We show that NFAT migration is exquisitely sensitive to Ca(2+) microdomains near open CRAC channels. Another Ca(2+)-permeable ion channel (TRPC3) was ineffective in driving NFAT1 to the nucleus. NFAT1 movement is temporally dissociated from the time course of the Ca(2+) signal and remains within the nucleus for 10 times longer than the duration of the trigger Ca(2+) signal. Kinetic analyses of each step linking CRAC channel activation to NFAT1 nuclear residency reveals that the rate-limiting step is transcription factor exit from the nucleus. The slow deactivation of NFAT provides a mechanism whereby Ca(2+)-dependent responses can be sustained despite the termination of the initial Ca(2+) signal and helps explain how gene expression in nonexcitable cells can continue after the primary stimulus has been removed.
Figures



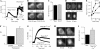

Similar articles
-
Distinct spatial Ca2+ signatures selectively activate different NFAT transcription factor isoforms.Mol Cell. 2015 Apr 16;58(2):232-43. doi: 10.1016/j.molcel.2015.02.027. Epub 2015 Mar 26. Mol Cell. 2015. PMID: 25818645 Free PMC article.
-
Signalling to transcription: store-operated Ca2+ entry and NFAT activation in lymphocytes.Cell Calcium. 2007 Aug;42(2):145-56. doi: 10.1016/j.ceca.2007.03.007. Epub 2007 Jun 18. Cell Calcium. 2007. PMID: 17572487 Review.
-
Ca(2+) Channel Re-localization to Plasma-Membrane Microdomains Strengthens Activation of Ca(2+)-Dependent Nuclear Gene Expression.Cell Rep. 2015 Jul 14;12(2):203-16. doi: 10.1016/j.celrep.2015.06.018. Epub 2015 Jul 2. Cell Rep. 2015. PMID: 26146085 Free PMC article.
-
Store-Operated Ca2+ Release-Activated Ca2+ Channels Regulate PAR2-Activated Ca2+ Signaling and Cytokine Production in Airway Epithelial Cells.J Immunol. 2015 Sep 1;195(5):2122-33. doi: 10.4049/jimmunol.1500396. Epub 2015 Aug 3. J Immunol. 2015. PMID: 26238490 Free PMC article.
-
The Role of Calcium-Calcineurin-NFAT Signaling Pathway in Health and Autoimmune Diseases.Front Immunol. 2020 Mar 10;11:195. doi: 10.3389/fimmu.2020.00195. eCollection 2020. Front Immunol. 2020. PMID: 32210952 Free PMC article. Review.
Cited by
-
A cholesterol-binding domain in STIM1 modulates STIM1-Orai1 physical and functional interactions.Sci Rep. 2016 Jul 27;6:29634. doi: 10.1038/srep29634. Sci Rep. 2016. PMID: 27459950 Free PMC article.
-
Endoplasmic reticulum-mitochondria coupling: local Ca²⁺ signalling with functional consequences.Pflugers Arch. 2012 Jul;464(1):27-32. doi: 10.1007/s00424-012-1095-x. Epub 2012 Mar 14. Pflugers Arch. 2012. PMID: 22415215 Review.
-
STIM proteins: dynamic calcium signal transducers.Nat Rev Mol Cell Biol. 2012 Sep;13(9):549-65. doi: 10.1038/nrm3414. Nat Rev Mol Cell Biol. 2012. PMID: 22914293 Free PMC article. Review.
-
Orai1-NFAT signalling pathway triggered by T cell receptor stimulation.Mol Cells. 2013 Mar;35(3):182-94. doi: 10.1007/s10059-013-0073-2. Epub 2013 Mar 11. Mol Cells. 2013. PMID: 23483280 Free PMC article. Review.
-
Multifaceted control of T cell differentiation by STIM1.Trends Biochem Sci. 2023 Dec;48(12):1083-1097. doi: 10.1016/j.tibs.2023.08.006. Epub 2023 Sep 9. Trends Biochem Sci. 2023. PMID: 37696713 Free PMC article. Review.
References
-
- Berridge M. J., Bootman M. D., Roderick H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529 - PubMed
-
- Neher E. (1998) Neuron 20, 389–399 - PubMed
-
- Rizzuto R., Pozzan T. (2006) Physiol. Rev. 86, 369–408 - PubMed
-
- Dolmetsch R. E., Pajvani U., Fife K., Spotts J. M., Greenberg M. E. (2001) Science 294, 333–339 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

