The hidden energetics of ligand binding and activation in a glutamate receptor
- PMID: 21317895
- PMCID: PMC3075596
- DOI: 10.1038/nsmb.2010
The hidden energetics of ligand binding and activation in a glutamate receptor
Abstract
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate most excitatory synaptic transmission in the central nervous system. The free energy of neurotransmitter binding to the ligand-binding domains (LBDs) of iGluRs is converted into useful work to drive receptor activation. We have computed the principal thermodynamic contributions from ligand docking and ligand-induced closure of LBDs for nine ligands of GluA2 using all-atom molecular dynamics free energy simulations. We have validated the results by comparison with experimentally measured apparent affinities to the isolated LBD. Features in the free energy landscapes that govern closure of LBDs are key determinants of binding free energies. An analysis of accessible LBD conformations transposed into the context of an intact GluA2 receptor revealed that the relative displacement of specific diagonal subunits in the tetrameric structure may be key to the action of partial agonists.
Figures

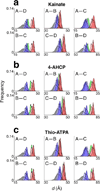
Comment in
-
Glutamate receptor ion channels: where do all the calories go?Nat Struct Mol Biol. 2011 Mar;18(3):253-4. doi: 10.1038/nsmb0311-253. Nat Struct Mol Biol. 2011. PMID: 21372852 No abstract available.
Similar articles
-
D-Serine Potently Drives Ligand-Binding Domain Closure in the Ionotropic Glutamate Receptor GluD2.Structure. 2020 Oct 6;28(10):1168-1178.e2. doi: 10.1016/j.str.2020.07.005. Epub 2020 Jul 30. Structure. 2020. PMID: 32735769 Free PMC article.
-
On the binding determinants of the glutamate agonist with the glutamate receptor ligand binding domain.Biochemistry. 2005 Aug 30;44(34):11508-17. doi: 10.1021/bi050547w. Biochemistry. 2005. PMID: 16114887
-
Retour aux sources: defining the structural basis of glutamate receptor activation.J Physiol. 2015 Jan 1;593(1):97-110. doi: 10.1113/jphysiol.2014.277921. Epub 2014 Oct 21. J Physiol. 2015. PMID: 25556791 Free PMC article. Review.
-
Structure of a glutamate-receptor ligand-binding core in complex with kainate.Nature. 1998 Oct 29;395(6705):913-7. doi: 10.1038/27692. Nature. 1998. PMID: 9804426
-
Emerging structural explanations of ionotropic glutamate receptor function.FASEB J. 2004 Mar;18(3):428-38. doi: 10.1096/fj.03-0873rev. FASEB J. 2004. PMID: 15003989 Review.
Cited by
-
Reduced curvature of ligand-binding domain free-energy surface underlies partial agonism at NMDA receptors.Structure. 2015 Jan 6;23(1):228-236. doi: 10.1016/j.str.2014.11.012. Epub 2014 Dec 24. Structure. 2015. PMID: 25543253 Free PMC article.
-
Functional insights from glutamate receptor ion channel structures.Annu Rev Physiol. 2013;75:313-37. doi: 10.1146/annurev-physiol-030212-183711. Epub 2012 Sep 4. Annu Rev Physiol. 2013. PMID: 22974439 Free PMC article. Review.
-
Blind prediction of charged ligand binding affinities in a model binding site.J Mol Biol. 2013 Nov 15;425(22):4569-83. doi: 10.1016/j.jmb.2013.07.030. Epub 2013 Jul 26. J Mol Biol. 2013. PMID: 23896298 Free PMC article.
-
Molecular lock regulates binding of glycine to a primitive NMDA receptor.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):E6786-E6795. doi: 10.1073/pnas.1607010113. Epub 2016 Oct 17. Proc Natl Acad Sci U S A. 2016. PMID: 27791085 Free PMC article.
-
Structure of an agonist-bound ionotropic glutamate receptor.Science. 2014 Aug 29;345(6200):1070-4. doi: 10.1126/science.1256508. Epub 2014 Aug 7. Science. 2014. PMID: 25103407 Free PMC article.
References
-
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. - PubMed
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
-
- Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

