Identification of PITX1 as a TERT suppressor gene located on human chromosome 5
- PMID: 21300782
- PMCID: PMC3126332
- DOI: 10.1128/MCB.00470-10
Identification of PITX1 as a TERT suppressor gene located on human chromosome 5
Abstract
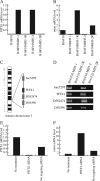
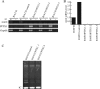
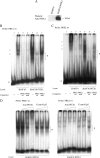
Telomerase, a ribonucleoprotein enzyme that maintains telomere length, is crucial for cellular immortalization and cancer progression. Telomerase activity is attributed primarily to the expression of telomerase reverse transcriptase (TERT). Using microcell-mediated chromosome transfer (MMCT) into the mouse melanoma cell line B16F10, we previously found that human chromosome 5 carries a gene, or genes, that can negatively regulate TERT expression (H. Kugoh, K. Shigenami, K. Funaki, J. Barrett, and M. Oshimura, Genes Chromosome Cancer 36:37-47, 2003). To identify the gene responsible for the regulation of TERT transcription, we performed cDNA microarray analysis using parental B16F10 cells, telomerase-negative B16F10 microcell hybrids with a human chromosome 5 (B16F10MH5), and its revertant clones (MH5R) with reactivated telomerase. Here, we report the identification of PITX1, whose expression leads to the downregulation of mouse tert (mtert) transcription, as a TERT suppressor gene. Additionally, both human TERT (hTERT) and mouse TERT (mtert) promoter activity can be suppressed by PITX1. We show that three and one binding site within the hTERT and mtert promoters, respectively, that express a unique conserved region are responsible for the transcriptional activation of TERT. Furthermore, we showed that PITX1 binds to the TERT promoter both in vitro and in vivo. Thus, PITX1 suppresses TERT transcription through direct binding to the TERT promoter, which ultimately regulates telomerase activity.
Figures




Similar articles
-
PITX1 protein interacts with ZCCHC10 to regulate hTERT mRNA transcription.PLoS One. 2019 Aug 12;14(8):e0217605. doi: 10.1371/journal.pone.0217605. eCollection 2019. PLoS One. 2019. PMID: 31404068 Free PMC article.
-
Human chromosome 5 carries a transcriptional regulator of human telomerase reverse transcriptase (hTERT).Biochem Biophys Res Commun. 2010 Aug 6;398(4):695-701. doi: 10.1016/j.bbrc.2010.07.003. Epub 2010 Jul 16. Biochem Biophys Res Commun. 2010. PMID: 20621064
-
miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells.Sci Rep. 2015 Feb 3;5:8201. doi: 10.1038/srep08201. Sci Rep. 2015. PMID: 25643913 Free PMC article.
-
Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms.Carcinogenesis. 2003 Jul;24(7):1167-76. doi: 10.1093/carcin/bgg085. Epub 2003 May 22. Carcinogenesis. 2003. PMID: 12807729 Review.
-
Telomerase regulation.Mutat Res. 2012 Feb 1;730(1-2):20-7. doi: 10.1016/j.mrfmmm.2011.10.003. Epub 2011 Oct 18. Mutat Res. 2012. PMID: 22032831 Free PMC article. Review.
Cited by
-
Regulation of the human catalytic subunit of telomerase (hTERT).Gene. 2012 May 1;498(2):135-46. doi: 10.1016/j.gene.2012.01.095. Epub 2012 Feb 13. Gene. 2012. PMID: 22381618 Free PMC article. Review.
-
PITX1 is a reliable biomarker for predicting prognosis in patients with oral epithelial dysplasia.Oncol Lett. 2014 Mar;7(3):750-754. doi: 10.3892/ol.2013.1775. Epub 2013 Dec 24. Oncol Lett. 2014. PMID: 24527083 Free PMC article.
-
Interaction between physical activity, PITX1 rs647161 genetic polymorphism and colorectal cancer risk in a Korean population: a case-control study.Oncotarget. 2018 Jan 10;9(7):7590-7603. doi: 10.18632/oncotarget.24136. eCollection 2018 Jan 26. Oncotarget. 2018. PMID: 29484135 Free PMC article.
-
Intragenic DNA methylation of PITX1 and the adjacent long non-coding RNA C5orf66-AS1 are prognostic biomarkers in patients with head and neck squamous cell carcinomas.PLoS One. 2018 Feb 9;13(2):e0192742. doi: 10.1371/journal.pone.0192742. eCollection 2018. PLoS One. 2018. PMID: 29425237 Free PMC article.
-
Identification of a Potential Regulatory Variant for Colorectal Cancer Risk Mapping to Chromosome 5q31.1: A Post-GWAS Study.PLoS One. 2015 Sep 18;10(9):e0138478. doi: 10.1371/journal.pone.0138478. eCollection 2015. PLoS One. 2015. PMID: 26381143 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
