A novel knowledge-driven systems biology approach for phenotype prediction upon genetic intervention
- PMID: 21282866
- PMCID: PMC3211072
- DOI: 10.1109/TCBB.2011.18
A novel knowledge-driven systems biology approach for phenotype prediction upon genetic intervention
Abstract
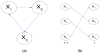
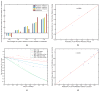
Deciphering the biological networks underlying complex phenotypic traits, e.g., human disease is undoubtedly crucial to understand the underlying molecular mechanisms and to develop effective therapeutics. Due to the network complexity and the relatively small number of available experiments, data-driven modeling is a great challenge for deducing the functions of genes/proteins in the network and in phenotype formation. We propose a novel knowledge-driven systems biology method that utilizes qualitative knowledge to construct a Dynamic Bayesian network (DBN) to represent the biological network underlying a specific phenotype. Edges in this network depict physical interactions between genes and/or proteins. A qualitative knowledge model first translates typical molecular interactions into constraints when resolving the DBN structure and parameters. Therefore, the uncertainty of the network is restricted to a subset of models which are consistent with the qualitative knowledge. All models satisfying the constraints are considered as candidates for the underlying network. These consistent models are used to perform quantitative inference. By in silico inference, we can predict phenotypic traits upon genetic interventions and perturbing in the network. We applied our method to analyze the puzzling mechanism of breast cancer cell proliferation network and we accurately predicted cancer cell growth rate upon manipulating (anti)cancerous marker genes/proteins.
Figures



Similar articles
-
Fitting Boolean networks from steady state perturbation data.Stat Appl Genet Mol Biol. 2011 Oct 5;10(1):47. doi: 10.2202/1544-6115.1727. Stat Appl Genet Mol Biol. 2011. PMID: 23089817 Free PMC article.
-
Network inference using informative priors.Proc Natl Acad Sci U S A. 2008 Sep 23;105(38):14313-8. doi: 10.1073/pnas.0802272105. Epub 2008 Sep 17. Proc Natl Acad Sci U S A. 2008. PMID: 18799736 Free PMC article.
-
Modularity-based credible prediction of disease genes and detection of disease subtypes on the phenotype-gene heterogeneous network.BMC Syst Biol. 2011 May 20;5:79. doi: 10.1186/1752-0509-5-79. BMC Syst Biol. 2011. PMID: 21599985 Free PMC article.
-
Causal Queries from Observational Data in Biological Systems via Bayesian Networks: An Empirical Study in Small Networks.Methods Mol Biol. 2019;1883:111-142. doi: 10.1007/978-1-4939-8882-2_5. Methods Mol Biol. 2019. PMID: 30547398 Review.
-
Systems pathology--taking molecular pathology into a new dimension.Nat Rev Clin Oncol. 2009 Aug;6(8):455-64. doi: 10.1038/nrclinonc.2009.102. Epub 2009 Jul 7. Nat Rev Clin Oncol. 2009. PMID: 19581910 Review.
Cited by
-
Predictive network modeling in human induced pluripotent stem cells identifies key driver genes for insulin responsiveness.PLoS Comput Biol. 2020 Dec 23;16(12):e1008491. doi: 10.1371/journal.pcbi.1008491. eCollection 2020 Dec. PLoS Comput Biol. 2020. PMID: 33362275 Free PMC article.
-
Changing Trends in Computational Drug Repositioning.Pharmaceuticals (Basel). 2018 Jun 5;11(2):57. doi: 10.3390/ph11020057. Pharmaceuticals (Basel). 2018. PMID: 29874824 Free PMC article. Review.
-
From integrative disease modeling to predictive, preventive, personalized and participatory (P4) medicine.EPMA J. 2013 Nov 6;4(1):23. doi: 10.1186/1878-5085-4-23. EPMA J. 2013. PMID: 24195840 Free PMC article.
-
A comprehensive review of artificial intelligence and network based approaches to drug repurposing in Covid-19.Biomed Pharmacother. 2022 Sep;153:113350. doi: 10.1016/j.biopha.2022.113350. Epub 2022 Jun 28. Biomed Pharmacother. 2022. PMID: 35777222 Free PMC article. Review.
-
Systems biology data analysis methodology in pharmacogenomics.Pharmacogenomics. 2011 Sep;12(9):1349-60. doi: 10.2217/pgs.11.76. Pharmacogenomics. 2011. PMID: 21919609 Free PMC article. Review.
References
-
- Bayes T. An Essay Towards Solving a Problem in the Doctrine of Chances. Philosophical Trans Royal Soc of London. 1763;53:370–418.
-
- Beal MJ, Falciani F, Ghahramani Z, Rangel C, Wild DL. A Bayesian Approach to Reconstructing Genetic Regulatory Networks with Hidden Factors. Bioinformatics. 2005;21:349–356. - PubMed
-
- Caldon CE, Daly RJ, Sutherland RL, Musgrove EA. Cell Cycle Control in Breast Cancer Cells. J Cellular Biochemistry. 2006;97(2):261–274. - PubMed
-
- Chang R. Consistent Modeling, Integration and Simulation of Molecular Interaction Networks in Space-Time Dimension. Proc. Seventh IEEE Int’l Conf. Bioinformatics and Bioengineering (BIBE ’07); 2007. pp. 1254–1259.
-
- Chang R, Brauer W, Stetter M. Modeling Semantics of Inconsistent Qualitative Knowledge for Quantitative Bayesian Network Inference. Neural Networks. 2008;21(2–3):182–192. - PubMed

