Replication protein A safeguards genome integrity by controlling NER incision events
- PMID: 21282463
- PMCID: PMC3101093
- DOI: 10.1083/jcb.201006011
Replication protein A safeguards genome integrity by controlling NER incision events
Abstract
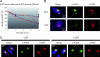
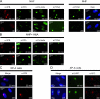
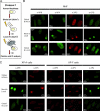
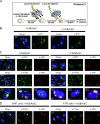
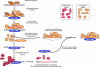
Single-stranded DNA gaps that might arise by futile repair processes can lead to mutagenic events and challenge genome integrity. Nucleotide excision repair (NER) is an evolutionarily conserved repair mechanism, essential for removal of helix-distorting DNA lesions. In the currently prevailing model, NER operates through coordinated assembly of repair factors into pre- and post-incision complexes; however, its regulation in vivo is poorly understood. Notably, the transition from dual incision to repair synthesis should be rigidly synchronized as it might lead to accumulation of unprocessed repair intermediates. We monitored NER regulatory events in vivo using sequential UV irradiations. Under conditions that allow incision yet prevent completion of repair synthesis or ligation, preincision factors can reassociate with new damage sites. In contrast, replication protein A remains at the incomplete NER sites and regulates a feedback loop from completion of DNA repair synthesis to subsequent damage recognition, independently of ATR signaling. Our data reveal an important function for replication protein A in averting further generation of DNA strand breaks that could lead to mutagenic and recombinogenic events.
Figures
Similar articles
-
DNA damage response and transcription.DNA Repair (Amst). 2011 Jul 15;10(7):743-50. doi: 10.1016/j.dnarep.2011.04.024. Epub 2011 May 31. DNA Repair (Amst). 2011. PMID: 21622031 Review.
-
Differential binding kinetics of replication protein A during replication and the pre- and post-incision steps of nucleotide excision repair.DNA Repair (Amst). 2014 Dec;24:46-56. doi: 10.1016/j.dnarep.2014.09.013. Epub 2014 Oct 16. DNA Repair (Amst). 2014. PMID: 25453469
-
Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells.J Cell Sci. 2007 Mar 15;120(Pt 6):1104-12. doi: 10.1242/jcs.03391. Epub 2007 Feb 27. J Cell Sci. 2007. PMID: 17327276
-
Gap-filling and bypass at the replication fork are both active mechanisms for tolerance of low-dose ultraviolet-induced DNA damage in the human genome.DNA Repair (Amst). 2014 Feb;14:27-38. doi: 10.1016/j.dnarep.2013.12.005. Epub 2013 Dec 28. DNA Repair (Amst). 2014. PMID: 24380689
-
Damage removal and gap filling in nucleotide excision repair.Enzymes. 2019;45:59-97. doi: 10.1016/bs.enz.2019.06.001. Epub 2019 Jul 13. Enzymes. 2019. PMID: 31627883 Review.
Cited by
-
Replication protein A prevents promiscuous annealing between short sequence homologies: Implications for genome integrity.Bioessays. 2015 Mar;37(3):305-13. doi: 10.1002/bies.201400161. Epub 2014 Nov 14. Bioessays. 2015. PMID: 25400143 Free PMC article. Review.
-
Integrated stress response control of granulosa cell translation and proliferation during normal ovarian follicle development.Mol Hum Reprod. 2021 Aug 7;27(8):gaab050. doi: 10.1093/molehr/gaab050. Mol Hum Reprod. 2021. PMID: 34314477 Free PMC article.
-
Impact of Age and Insulin-Like Growth Factor-1 on DNA Damage Responses in UV-Irradiated Human Skin.Molecules. 2017 Feb 26;22(3):356. doi: 10.3390/molecules22030356. Molecules. 2017. PMID: 28245638 Free PMC article. Review.
-
XPA: DNA Repair Protein of Significant Clinical Importance.Int J Mol Sci. 2020 Mar 22;21(6):2182. doi: 10.3390/ijms21062182. Int J Mol Sci. 2020. PMID: 32235701 Free PMC article. Review.
-
RPA and XPA interaction with DNA structures mimicking intermediates of the late stages in nucleotide excision repair.PLoS One. 2018 Jan 10;13(1):e0190782. doi: 10.1371/journal.pone.0190782. eCollection 2018. PLoS One. 2018. PMID: 29320546 Free PMC article.
References
-
- Araújo S.J., Tirode F., Coin F., Pospiech H., Syväoja J.E., Stucki M., Hübscher U., Egly J.M., Wood R.D. 2000. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14:349–359 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

