PiNGO: a Cytoscape plugin to find candidate genes in biological networks
- PMID: 21278188
- PMCID: PMC3065683
- DOI: 10.1093/bioinformatics/btr045
PiNGO: a Cytoscape plugin to find candidate genes in biological networks
Abstract
PiNGO is a tool to screen biological networks for candidate genes, i.e. genes predicted to be involved in a biological process of interest. The user can narrow the search to genes with particular known functions or exclude genes belonging to particular functional classes. PiNGO provides support for a wide range of organisms and Gene Ontology classification schemes, and it can easily be customized for other organisms and functional classifications. PiNGO is implemented as a plugin for Cytoscape, a popular network visualization platform.
Availability: PiNGO is distributed as an open-source Java package under the GNU General Public License (http://www.gnu.org/), and can be downloaded via the Cytoscape plugin manager. A detailed user guide and tutorial are available on the PiNGO website (http://www.psb.ugent.be/esb/PiNGO.
Figures
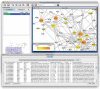
Similar articles
-
GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop.Bioinformatics. 2010 Nov 15;26(22):2927-8. doi: 10.1093/bioinformatics/btq562. Epub 2010 Oct 5. Bioinformatics. 2010. PMID: 20926419 Free PMC article.
-
NOA: a cytoscape plugin for network ontology analysis.Bioinformatics. 2013 Aug 15;29(16):2066-7. doi: 10.1093/bioinformatics/btt334. Epub 2013 Jun 7. Bioinformatics. 2013. PMID: 23749961 Free PMC article.
-
Mosaic: making biological sense of complex networks.Bioinformatics. 2012 Jul 15;28(14):1943-4. doi: 10.1093/bioinformatics/bts278. Epub 2012 May 9. Bioinformatics. 2012. PMID: 22576176 Free PMC article.
-
JEPETTO: a Cytoscape plugin for gene set enrichment and topological analysis based on interaction networks.Bioinformatics. 2014 Apr 1;30(7):1029-30. doi: 10.1093/bioinformatics/btt732. Epub 2013 Dec 19. Bioinformatics. 2014. PMID: 24363376 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
Cited by
-
Transcriptome analysis of lipid biosynthesis during kernel development in two walnut (Juglans regia L.) varieties of 'Xilin 3' and 'Xiangling'.BMC Plant Biol. 2024 Sep 4;24(1):828. doi: 10.1186/s12870-024-05546-y. BMC Plant Biol. 2024. PMID: 39227757 Free PMC article.
-
Identifying gene interaction networks.Methods Mol Biol. 2012;850:483-94. doi: 10.1007/978-1-61779-555-8_26. Methods Mol Biol. 2012. PMID: 22307715 Free PMC article.
-
MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours.Br J Cancer. 2016 Apr 12;114(8):917-28. doi: 10.1038/bjc.2016.46. Epub 2016 Mar 8. Br J Cancer. 2016. PMID: 26954716 Free PMC article.
-
Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants.Plant Cell. 2013 Aug;25(8):2865-77. doi: 10.1105/tpc.113.112268. Epub 2013 Aug 13. Plant Cell. 2013. PMID: 23943861 Free PMC article.
-
RNA Sequencing and Coexpression Analysis Reveal Key Genes Involved in α-Linolenic Acid Biosynthesis in Perilla frutescens Seed.Int J Mol Sci. 2017 Nov 16;18(11):2433. doi: 10.3390/ijms18112433. Int J Mol Sci. 2017. PMID: 29144390 Free PMC article.
References
-
- Aerts S., et al. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24:537–544. - PubMed
-
- Chua H.N., et al. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics. 2006;22:1623–1630. - PubMed
-
- Hughes T.R., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. - PubMed
-
- Murali T.M., et al. The art of gene function prediction. Nat. Biotechnol. 2006;24:1474–1475. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

