LEDGF dominant interference proteins demonstrate prenuclear exposure of HIV-1 integrase and synergize with LEDGF depletion to destroy viral infectivity
- PMID: 21270171
- PMCID: PMC3067863
- DOI: 10.1128/JVI.01295-10
LEDGF dominant interference proteins demonstrate prenuclear exposure of HIV-1 integrase and synergize with LEDGF depletion to destroy viral infectivity
Abstract
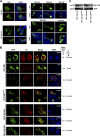
Target cell overexpression of the integrase binding domain (IBD) of LEDGF/p75 (LEDGF) inhibits HIV-1 replication. The mechanism and protein structure requirements for this dominant interference are unclear. More generally, how and when HIV-1 uncoating occurs postentry is poorly defined, and it is unknown whether integrase within the evolving viral core becomes accessible to cellular proteins prior to nuclear entry. We used LEDGF dominant interference to address the latter question while characterizing determinants of IBD antiviral activity. Fusions of green fluorescent protein (GFP) with multiple C-terminal segments of LEDGF inhibited HIV-1 replication substantially, but minimal chimeras of either polarity (GFP-IBD or IBD-GFP) were most effective. Combining GFP-IBD expression with LEDGF depletion was profoundly antiviral. CD4(+) T cell lines were rendered virtually uninfectable, with single-cycle HIV-1 infectivity reduced 4 logs and high-input (multiplicity of infection = 5.0) replication completely blocked. We restricted GFP-IBD to specific intracellular locations and found that antiviral activity was preserved when the protein was confined to the cytoplasm or directed to the nuclear envelope. The life cycle block triggered by the cytoplasm-restricted protein manifested after nuclear entry, at the level of integration. We conclude that integrase within the viral core becomes accessible to host cell protein interaction in the cytoplasm. LEDGF dominant interference and depletion impair HIV-1 integration at distinct postentry stages. GFP-IBD may trigger premature or improper integrase oligomerization.
Figures

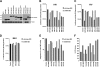
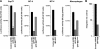

Similar articles
-
LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors.PLoS Pathog. 2009 Jul;5(7):e1000522. doi: 10.1371/journal.ppat.1000522. Epub 2009 Jul 17. PLoS Pathog. 2009. PMID: 19609362 Free PMC article.
-
TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism.J Virol. 2014 Sep 1;88(17):9704-17. doi: 10.1128/JVI.01397-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942577 Free PMC article.
-
LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs.PLoS Pathog. 2012;8(3):e1002558. doi: 10.1371/journal.ppat.1002558. Epub 2012 Mar 1. PLoS Pathog. 2012. PMID: 22396646 Free PMC article.
-
The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy.Virology. 2013 Jan 5;435(1):102-9. doi: 10.1016/j.virol.2012.09.033. Virology. 2013. PMID: 23217620 Review.
-
[LEDGF/p75: a novel target for anti-HIV therapy and advances in the study of its related inhibitors].Yao Xue Xue Bao. 2009 Sep;44(9):953-60. Yao Xue Xue Bao. 2009. PMID: 20055168 Review. Chinese.
Cited by
-
HIV-1 Preintegration Complex Preferentially Integrates the Viral DNA into Nucleosomes Containing Trimethylated Histone 3-Lysine 36 Modification and Flanking Linker DNA.J Virol. 2022 Sep 28;96(18):e0101122. doi: 10.1128/jvi.01011-22. Epub 2022 Sep 12. J Virol. 2022. PMID: 36094316 Free PMC article.
-
HIV-1 Activation of Innate Immunity Depends Strongly on the Intracellular Level of TREX1 and Sensing of Incomplete Reverse Transcription Products.J Virol. 2018 Jul 31;92(16):e00001-18. doi: 10.1128/JVI.00001-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29769349 Free PMC article.
-
Retroviral DNA Integration.Chem Rev. 2016 Oct 26;116(20):12730-12757. doi: 10.1021/acs.chemrev.6b00125. Epub 2016 May 20. Chem Rev. 2016. PMID: 27198982 Free PMC article. Review.
-
LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions.Retrovirology. 2013 May 30;10:57. doi: 10.1186/1742-4690-10-57. Retrovirology. 2013. PMID: 23721378 Free PMC article.
-
Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes.Viruses. 2013 Oct 7;5(10):2483-511. doi: 10.3390/v5102483. Viruses. 2013. PMID: 24103892 Free PMC article. Review.
References
-
- Bao, K. K., et al. 2003. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 278:1323-1327. - PubMed
-
- Bartholomeeusen, K., et al. 2007. Differential interaction of HIV-1 integrase and JPO2 with the C terminus of LEDGF/p75. J. Mol. Biol. 372:407-421. - PubMed
-
- Busschots, K., et al. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

