Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop
- PMID: 21270156
- PMCID: PMC3067856
- DOI: 10.1128/JVI.02658-10
Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop
Abstract
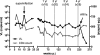
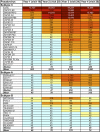
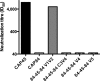
The targets of broadly cross-neutralizing (BCN) antibodies are of great interest in the HIV vaccine field. We have identified a subtype C HIV-1-superinfected individual, CAP256, with high-level BCN activity, and characterized the antibody specificity mediating breadth. CAP256 developed potent BCN activity peaking at 3 years postinfection, neutralizing 32 (76%) of 42 heterologous viruses, with titers of antibodies against some viruses exceeding 1:10,000. CAP256 showed a subtype bias, preferentially neutralizing subtype C and A viruses over subtype B viruses. CAP256 BCN serum targeted a quaternary epitope which included the V1V2 region. Further mapping identified residues F159, N160, L165, R166, D167, K169, and K171 (forming the FN/LRD-K-K motif) in the V2 region as crucial to the CAP256 epitope. However, the fine specificity of the BCN response varied over time and, while consistently dependent on R166 and K169, became gradually less dependent on D167 and K171, possibly contributing to the incremental increase in breadth over 4 years. The presence of an intact FN/LRD-K-K motif in heterologous viruses was associated with sensitivity, although the length of the adjacent V1 loop modulated the degree of sensitivity, with a shorter V1 region significantly associated with higher titers. Repair of the FN/LRD-K-K motif in resistant heterologous viruses conferred sensitivity, with titers sometimes exceeding 1:10,000. Comparison of the CAP256 epitope with that of the PG9/PG16 monoclonal antibodies suggested that these epitopes overlapped, adding to the mounting evidence that this may represent a common neutralization target that should be further investigated as a potential vaccine candidate.
Figures
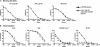





Similar articles
-
Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies.J Virol. 2013 May;87(9):4882-94. doi: 10.1128/JVI.03424-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408621 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency.J Virol. 2015 Oct 14;90(1):76-91. doi: 10.1128/JVI.01791-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468542 Free PMC article.
-
The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection.J Virol. 2011 May;85(10):4828-40. doi: 10.1128/JVI.00198-11. Epub 2011 Mar 9. J Virol. 2011. PMID: 21389135 Free PMC article.
-
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.J Virol. 2018 Feb 12;92(5):e01796-17. doi: 10.1128/JVI.01796-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237847 Free PMC article.
Cited by
-
An efficiently cleaved HIV-1 clade C Env selectively binds to neutralizing antibodies.PLoS One. 2015 Mar 30;10(3):e0122443. doi: 10.1371/journal.pone.0122443. eCollection 2015. PLoS One. 2015. PMID: 25822521 Free PMC article.
-
Neutralizing antibody escape during HIV-1 mother-to-child transmission involves conformational masking of distal epitopes in envelope.J Virol. 2012 Sep;86(18):9566-82. doi: 10.1128/JVI.00953-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740394 Free PMC article.
-
Virological features associated with the development of broadly neutralizing antibodies to HIV-1.Trends Microbiol. 2015 Apr;23(4):204-11. doi: 10.1016/j.tim.2014.12.007. Epub 2015 Jan 5. Trends Microbiol. 2015. PMID: 25572881 Free PMC article. Review.
-
Anti-V3/Glycan and Anti-MPER Neutralizing Antibodies, but Not Anti-V2/Glycan Site Antibodies, Are Strongly Associated with Greater Anti-HIV-1 Neutralization Breadth and Potency.J Virol. 2015 May;89(10):5264-75. doi: 10.1128/JVI.00129-15. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673728 Free PMC article.
-
Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site.J Virol. 2012 May;86(10):5844-56. doi: 10.1128/JVI.07139-11. Epub 2012 Mar 14. J Virol. 2012. PMID: 22419808 Free PMC article.
References
-
- Brown, B. K., et al. 2008. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 375:529-538. - PubMed
-
- Burton, D. R., et al. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

