Chatting histone modifications in mammals
- PMID: 21266346
- PMCID: PMC3080777
- DOI: 10.1093/bfgp/elq024
Chatting histone modifications in mammals
Abstract
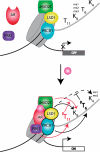
Eukaryotic chromatin can be highly dynamic and can continuously exchange between an open transcriptionally active conformation and a compacted silenced one. Post-translational modifications of histones have a pivotal role in regulating chromatin states, thus influencing all chromatin dependent processes. Methylation is currently one of the best characterized histone modification and occurs on arginine and lysine residues. Histone methylation can regulate other modifications (e.g. acetylation, phosphorylation and ubiquitination) in order to define a precise functional chromatin environment. In this review we focus on histone methylation and demethylation, as well as on the enzymes responsible for setting these marks. In particular we are describing novel concepts on the interdependence of histone modifications marks and discussing the molecular mechanisms governing this cross-talks.
Figures

Similar articles
-
Histone lysine methylation and chromatin replication.Biochim Biophys Acta. 2014 Dec;1839(12):1433-9. doi: 10.1016/j.bbagrm.2014.03.009. Epub 2014 Mar 28. Biochim Biophys Acta. 2014. PMID: 24686120 Review.
-
Chromatin dynamics: interplay between remodeling enzymes and histone modifications.Biochim Biophys Acta. 2014 Aug;1839(8):728-36. doi: 10.1016/j.bbagrm.2014.02.013. Epub 2014 Feb 28. Biochim Biophys Acta. 2014. PMID: 24583555 Free PMC article. Review.
-
Histone H3 phosphorylation - a versatile chromatin modification for different occasions.Biochimie. 2012 Nov;94(11):2193-201. doi: 10.1016/j.biochi.2012.04.018. Epub 2012 Apr 28. Biochimie. 2012. PMID: 22564826 Free PMC article. Review.
-
Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation.Curr Opin Cell Biol. 2008 Jun;20(3):341-8. doi: 10.1016/j.ceb.2008.03.019. Epub 2008 May 26. Curr Opin Cell Biol. 2008. PMID: 18508253 Free PMC article. Review.
-
Histone crosstalk directed by H2B ubiquitination is required for chromatin boundary integrity.PLoS Genet. 2011 Jul;7(7):e1002175. doi: 10.1371/journal.pgen.1002175. Epub 2011 Jul 21. PLoS Genet. 2011. PMID: 21811414 Free PMC article.
Cited by
-
Cohesin codes - interpreting chromatin architecture and the many facets of cohesin function.J Cell Sci. 2013 Jan 1;126(Pt 1):31-41. doi: 10.1242/jcs.116566. J Cell Sci. 2013. PMID: 23516328 Free PMC article. Review.
-
Regulation of DU145 prostate cancer cell growth by Scm-like with four mbt domains 2.J Biosci. 2013 Mar;38(1):105-12. doi: 10.1007/s12038-012-9283-6. J Biosci. 2013. PMID: 23385818
-
Histones and their modifications in ovarian cancer - drivers of disease and therapeutic targets.Front Oncol. 2014 Jun 12;4:144. doi: 10.3389/fonc.2014.00144. eCollection 2014. Front Oncol. 2014. PMID: 24971229 Free PMC article. Review.
-
Glucocorticoids regulate natural killer cell function epigenetically.Cell Immunol. 2014 Jul;290(1):120-30. doi: 10.1016/j.cellimm.2014.05.013. Epub 2014 Jun 18. Cell Immunol. 2014. PMID: 24978612 Free PMC article.
-
HDAC8 substrate selectivity is determined by long- and short-range interactions leading to enhanced reactivity for full-length histone substrates compared with peptides.J Biol Chem. 2017 Dec 29;292(52):21568-21577. doi: 10.1074/jbc.M117.811026. Epub 2017 Nov 6. J Biol Chem. 2017. PMID: 29109148 Free PMC article.
References
-
- Luger K, Rechsteiner TJ, Flaus AJ, et al. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272(3):301–11. - PubMed
-
- Kunert N, Brehm A. Novel Mi-2 related ATP-dependent chromatin remodelers. Epigenetics. 2009;4(4):209–11. - PubMed
-
- Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11(4):264–75. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. - PubMed
-
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

