Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast
- PMID: 21263029
- PMCID: PMC3172170
- DOI: 10.1083/jcb.201007018
Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast
Abstract
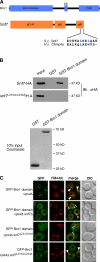
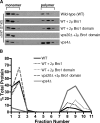
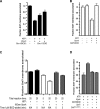
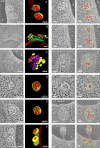
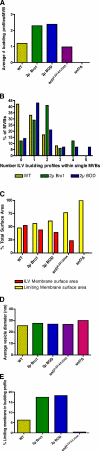
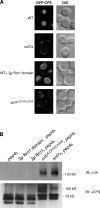
Endosomal sorting complexes required for transport (ESCRTs) promote the invagination of vesicles into the lumen of endosomes, the budding of enveloped viruses, and the separation of cells during cytokinesis. These processes share a topologically similar membrane scission event facilitated by ESCRT-III assembly at the cytosolic surface of the membrane. The Snf7 subunit of ESCRT-III in yeast binds directly to an auxiliary protein, Bro1. Like ESCRT-III, Bro1 is required for the formation of intralumenal vesicles at endosomes, but its role in membrane scission is unknown. We show that overexpression of Bro1 or its N-terminal Bro1 domain that binds Snf7 enhances the stability of ESCRT-III by inhibiting Vps4-mediated disassembly in vivo and in vitro. This stabilization effect correlates with a reduced frequency in the detachment of intralumenal vesicles as observed by electron tomography, implicating Bro1 as a regulator of ESCRT-III disassembly and membrane scission activity.
Figures
Similar articles
-
Interaction maps of the Saccharomyces cerevisiae ESCRT-III protein Snf7.Eukaryot Cell. 2013 Nov;12(11):1538-46. doi: 10.1128/EC.00241-13. Epub 2013 Sep 20. Eukaryot Cell. 2013. PMID: 24058170 Free PMC article.
-
Membrane scission by the ESCRT-III complex.Nature. 2009 Mar 12;458(7235):172-7. doi: 10.1038/nature07836. Epub 2009 Feb 22. Nature. 2009. PMID: 19234443 Free PMC article.
-
Bro1 binds the Vps20 subunit of ESCRT-III and promotes ESCRT-III regulation by Doa4.Traffic. 2022 Feb;23(2):109-119. doi: 10.1111/tra.12828. Epub 2022 Jan 13. Traffic. 2022. PMID: 34908216 Free PMC article.
-
Membrane budding and scission by the ESCRT machinery: it's all in the neck.Nat Rev Mol Cell Biol. 2010 Aug;11(8):556-66. doi: 10.1038/nrm2937. Epub 2010 Jun 30. Nat Rev Mol Cell Biol. 2010. PMID: 20588296 Free PMC article. Review.
-
ALIX and the multivesicular endosome: ALIX in Wonderland.Trends Cell Biol. 2014 Jan;24(1):19-25. doi: 10.1016/j.tcb.2013.10.009. Epub 2013 Nov 26. Trends Cell Biol. 2014. PMID: 24287454 Review.
Cited by
-
Life in the lumen: The multivesicular endosome.Traffic. 2020 Jan;21(1):76-93. doi: 10.1111/tra.12715. Traffic. 2020. PMID: 31854087 Free PMC article. Review.
-
A plant Bro1 domain protein BRAF regulates multivesicular body biogenesis and membrane protein homeostasis.Nat Commun. 2018 Sep 17;9(1):3784. doi: 10.1038/s41467-018-05913-y. Nat Commun. 2018. PMID: 30224707 Free PMC article.
-
The ubiquitin hydrolase Doa4 directly binds Snf7 to inhibit recruitment of ESCRT-III remodeling factors in S. cerevisiae.J Cell Sci. 2020 Apr 28;133(8):jcs241455. doi: 10.1242/jcs.241455. J Cell Sci. 2020. PMID: 32184262 Free PMC article.
-
Three-dimensional architecture of ESCRT-III flat spirals on the membrane.Proc Natl Acad Sci U S A. 2024 May 14;121(20):e2319115121. doi: 10.1073/pnas.2319115121. Epub 2024 May 6. Proc Natl Acad Sci U S A. 2024. PMID: 38709931 Free PMC article.
-
Membrane dynamics during cytokinesis.Curr Opin Cell Biol. 2013 Feb;25(1):92-8. doi: 10.1016/j.ceb.2012.10.012. Epub 2012 Nov 21. Curr Opin Cell Biol. 2013. PMID: 23177492 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

