Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins
- PMID: 21261285
- PMCID: PMC3050080
- DOI: 10.1021/bi102057m
Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins
Abstract
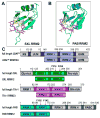
The RNA recognition motif (RRM) is a prevalent class of RNA binding domains. Although a number of RRM/RNA structures have been determined, thermodynamic analyses are relatively uncommon. Here, we use isothermal titration calorimetry to characterize single-stranded (ss)RNA binding by four representative RRM-containing proteins: (i) U2AF(65), (ii) SXL, (iii) TIA-1, and (iv) PAB. In all cases, ssRNA binding is accompanied by remarkably large favorable enthalpy changes (-30 to -60 kcal mol(-1)) and unfavorable entropy changes. Alterations of key RRM residues and binding sites indicate that under the nearly physiological conditions of these studies, large thermodynamic changes represent a signature of specific ssRNA recognition by RRMs.
Figures

Similar articles
-
Comparative analyses of the thermodynamic RNA binding signatures of different types of RNA recognition motifs.Nucleic Acids Res. 2017 Jun 2;45(10):6037-6050. doi: 10.1093/nar/gkx136. Nucleic Acids Res. 2017. PMID: 28334819 Free PMC article.
-
Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities.J Biol Chem. 1996 Feb 2;271(5):2783-8. doi: 10.1074/jbc.271.5.2783. J Biol Chem. 1996. PMID: 8576255
-
RRM-RNA recognition: NMR or crystallography…and new findings.Curr Opin Struct Biol. 2013 Feb;23(1):100-8. doi: 10.1016/j.sbi.2012.11.006. Epub 2012 Dec 14. Curr Opin Struct Biol. 2013. PMID: 23253355 Review.
-
Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1.J Mol Biol. 2012 Jan 27;415(4):727-40. doi: 10.1016/j.jmb.2011.11.040. Epub 2011 Dec 2. J Mol Biol. 2012. PMID: 22154808 Free PMC article.
-
U2AF homology motifs: protein recognition in the RRM world.Genes Dev. 2004 Jul 1;18(13):1513-26. doi: 10.1101/gad.1206204. Genes Dev. 2004. PMID: 15231733 Free PMC article. Review.
Cited by
-
The structure and selectivity of the SR protein SRSF2 RRM domain with RNA.Nucleic Acids Res. 2012 Apr;40(7):3232-44. doi: 10.1093/nar/gkr1164. Epub 2011 Dec 2. Nucleic Acids Res. 2012. PMID: 22140111 Free PMC article.
-
A Structure-free Method for Quantifying Conformational Flexibility in proteins.Sci Rep. 2016 Jun 30;6:29040. doi: 10.1038/srep29040. Sci Rep. 2016. PMID: 27358108 Free PMC article.
-
Comparative analyses of the thermodynamic RNA binding signatures of different types of RNA recognition motifs.Nucleic Acids Res. 2017 Jun 2;45(10):6037-6050. doi: 10.1093/nar/gkx136. Nucleic Acids Res. 2017. PMID: 28334819 Free PMC article.
-
Thermodynamic studies of a series of homologous HIV-1 TAR RNA ligands reveal that loose binders are stronger Tat competitors than tight ones.Nucleic Acids Res. 2013 Jun;41(11):5851-63. doi: 10.1093/nar/gkt237. Epub 2013 Apr 19. Nucleic Acids Res. 2013. PMID: 23605042 Free PMC article.
-
Kinked β-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL.Proc Natl Acad Sci U S A. 2011 Nov 8;108(45):18266-71. doi: 10.1073/pnas.1105211108. Epub 2011 Oct 20. Proc Natl Acad Sci U S A. 2011. PMID: 22021443 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

