Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria
- PMID: 21248725
- PMCID: PMC3118861
- DOI: 10.1038/mi.2010.83
Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria
Abstract
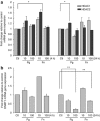
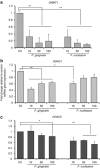
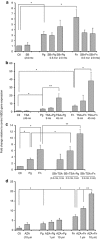
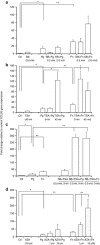
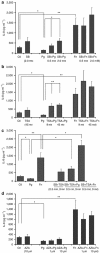
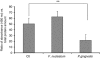
Gingival epithelia utilize multiple signaling pathways to regulate innate immune responses to various oral bacteria, but little is understood about how these bacteria alter epithelial epigenetic status. In this study we report that DNA methyltransferase (DNMT1) and histone deacetylase expression were decreased in gingival epithelial cells treated with oral pathogen Porphyromonas gingivalis and nonpathogen Fusobacterium nucleatum. Pretreatment with trichostatin A and sodium butyrate, which increase acetylation of chromatin histones, significantly enhanced the gene expression of antimicrobial proteins human β-defensin 2 (hBD2) and CC chemokine ligand 20 (CCL20) in response to both bacterial challenges. Pretreatment with DNMT inhibitor 5'-azacytidine increased hBD2 and CCL20 expression in response to F. nucleatum, but not to P. gingivalis. Furthermore, we observed a differential pattern of protein levels of H3K4me3, which has been associated with chromatin remodeling and activation of gene transcription, in response to P. gingivalis vs. F. nucleatum. This study provides a new insight into the bacteria-specific innate immune responses via epigenetic regulation.
Figures

Similar articles
-
Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier.Infect Immun. 2000 May;68(5):2907-15. doi: 10.1128/IAI.68.5.2907-2915.2000. Infect Immun. 2000. PMID: 10768988 Free PMC article.
-
Interplay of protease-activated receptors and NOD pattern recognition receptors in epithelial innate immune responses to bacteria.Immunol Lett. 2010 Jul 8;131(2):113-9. doi: 10.1016/j.imlet.2010.02.006. Epub 2010 Feb 26. Immunol Lett. 2010. PMID: 20219537 Free PMC article.
-
Immunologic characteristics of human gingival fibroblasts in response to oral bacteria.J Periodontal Res. 2017 Jun;52(3):447-457. doi: 10.1111/jre.12410. Epub 2016 Aug 24. J Periodontal Res. 2017. PMID: 27558278
-
Epigenetic regulation of inflammation in periodontitis: cellular mechanisms and therapeutic potential.Clin Epigenetics. 2020 Nov 30;12(1):186. doi: 10.1186/s13148-020-00982-7. Clin Epigenetics. 2020. PMID: 33256844 Free PMC article. Review.
-
Immunological Pathways Triggered by Porphyromonas gingivalis and Fusobacterium nucleatum: Therapeutic Possibilities?Mediators Inflamm. 2019 Jun 24;2019:7241312. doi: 10.1155/2019/7241312. eCollection 2019. Mediators Inflamm. 2019. PMID: 31341421 Free PMC article. Review.
Cited by
-
Epigenetics: the fine-tuner in inflammatory bowel disease?Curr Opin Gastroenterol. 2013 Jul;29(4):370-7. doi: 10.1097/MOG.0b013e328360bd12. Curr Opin Gastroenterol. 2013. PMID: 23743674 Free PMC article. Review.
-
Identification of a periodontal pathogen and bihormonal cells in pancreatic islets of humans and a mouse model of periodontitis.Sci Rep. 2020 Jun 19;10(1):9976. doi: 10.1038/s41598-020-65828-x. Sci Rep. 2020. PMID: 32561770 Free PMC article.
-
Modulation of gene transcription and epigenetics of colon carcinoma cells by bacterial membrane vesicles.Sci Rep. 2018 May 9;8(1):7434. doi: 10.1038/s41598-018-25308-9. Sci Rep. 2018. PMID: 29743643 Free PMC article.
-
Treponema denticola increases MMP-2 expression and activation in the periodontium via reversible DNA and histone modifications.Cell Microbiol. 2018 Apr;20(4):10.1111/cmi.12815. doi: 10.1111/cmi.12815. Epub 2018 Jan 8. Cell Microbiol. 2018. PMID: 29205773 Free PMC article.
-
Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics.Gastroenterology. 2013 Aug;145(2):293-308. doi: 10.1053/j.gastro.2013.05.050. Epub 2013 Jun 8. Gastroenterology. 2013. PMID: 23751777 Free PMC article. Review.
References
-
- Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429,:457–463. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128,:693–705. - PubMed
-
- Socransky S.S., Haffajee A.D., Cugini M.A., Smith C., Kent R.L., Jr Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25,:134–144. - PubMed
-
- Kinane D.F., Hart T.C. Genes and gene polymorphisms associated with periodontal disease. Crit. Rev. Oral. Biol. Med. 2003;14,:430–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

