Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation
- PMID: 21235772
- PMCID: PMC3025958
- DOI: 10.1186/1471-2164-12-36
Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation
Abstract
Background: Estrogen receptors alpha (ERα) and beta (ERβ) are transcription factors (TFs) that mediate estrogen signaling and define the hormone-responsive phenotype of breast cancer (BC). The two receptors can be found co-expressed and play specific, often opposite, roles, with ERβ being able to modulate the effects of ERα on gene transcription and cell proliferation. ERβ is frequently lost in BC, where its presence generally correlates with a better prognosis of the disease. The identification of the genomic targets of ERβ in hormone-responsive BC cells is thus a critical step to elucidate the roles of this receptor in estrogen signaling and tumor cell biology.
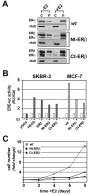
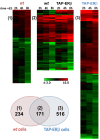
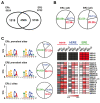
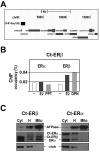
Results: Expression of full-length ERβ in hormone-responsive, ERα-positive MCF-7 cells resulted in a marked reduction in cell proliferation in response to estrogen and marked effects on the cell transcriptome. By ChIP-Seq we identified 9702 ERβ and 6024 ERα binding sites in estrogen-stimulated cells, comprising sites occupied by either ERβ, ERα or both ER subtypes. A search for TF binding matrices revealed that the majority of the binding sites identified comprise one or more Estrogen Response Element and the remaining show binding matrixes for other TFs known to mediate ER interaction with chromatin by tethering, including AP2, E2F and SP1. Of 921 genes differentially regulated by estrogen in ERβ+ vs ERβ- cells, 424 showed one or more ERβ site within 10 kb. These putative primary ERβ target genes control cell proliferation, death, differentiation, motility and adhesion, signal transduction and transcription, key cellular processes that might explain the biological and clinical phenotype of tumors expressing this ER subtype. ERβ binding in close proximity of several miRNA genes and in the mitochondrial genome, suggests the possible involvement of this receptor in small non-coding RNA biogenesis and mitochondrial genome functions.
Conclusions: Results indicate that the vast majority of the genomic targets of ERβ can bind also ERα, suggesting that the overall action of ERβ on the genome of hormone-responsive BC cells depends mainly on the relative concentration of both ERs in the cell.
Figures

Similar articles
-
Estrogen Receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding.Mol Endocrinol. 2008 May;22(5):1032-43. doi: 10.1210/me.2007-0356. Epub 2008 Feb 7. Mol Endocrinol. 2008. PMID: 18258689 Free PMC article.
-
Genome-wide dynamics of chromatin binding of estrogen receptors alpha and beta: mutual restriction and competitive site selection.Mol Endocrinol. 2010 Jan;24(1):47-59. doi: 10.1210/me.2009-0252. Epub 2009 Nov 6. Mol Endocrinol. 2010. PMID: 19897598 Free PMC article.
-
Direct regulation of microRNA biogenesis and expression by estrogen receptor beta in hormone-responsive breast cancer.Oncogene. 2012 Sep 20;31(38):4196-206. doi: 10.1038/onc.2011.583. Epub 2012 Jan 9. Oncogene. 2012. PMID: 22231442
-
ERbeta in breast cancer--onlooker, passive player, or active protector?Steroids. 2008 Oct;73(11):1039-51. doi: 10.1016/j.steroids.2008.04.006. Epub 2008 Apr 20. Steroids. 2008. PMID: 18501937 Free PMC article. Review.
-
Estrogen receptor alpha negative breast cancer patients: estrogen receptor beta as a therapeutic target.J Steroid Biochem Mol Biol. 2008 Mar;109(1-2):1-10. doi: 10.1016/j.jsbmb.2007.12.010. Epub 2007 Dec 8. J Steroid Biochem Mol Biol. 2008. PMID: 18243688 Review.
Cited by
-
Ancient Out-of-Africa Mitochondrial DNA Variants Associate with Distinct Mitochondrial Gene Expression Patterns.PLoS Genet. 2016 Nov 3;12(11):e1006407. doi: 10.1371/journal.pgen.1006407. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27812116 Free PMC article.
-
Estrogen Induces Selective Transcription of Caveolin1 Variants in Human Breast Cancer through Estrogen Responsive Element-Dependent Mechanisms.Int J Mol Sci. 2020 Aug 20;21(17):5989. doi: 10.3390/ijms21175989. Int J Mol Sci. 2020. PMID: 32825330 Free PMC article.
-
Identification of estrogen receptor dimer selective ligands reveals growth-inhibitory effects on cells that co-express ERα and ERβ.PLoS One. 2012;7(2):e30993. doi: 10.1371/journal.pone.0030993. Epub 2012 Feb 7. PLoS One. 2012. PMID: 22347418 Free PMC article.
-
Obesity as a Risk Factor for Breast Cancer-The Role of miRNA.Int J Mol Sci. 2022 Dec 10;23(24):15683. doi: 10.3390/ijms232415683. Int J Mol Sci. 2022. PMID: 36555323 Free PMC article. Review.
-
The genomic landscape of estrogen receptor α binding sites in mouse mammary gland.PLoS One. 2019 Aug 13;14(8):e0220311. doi: 10.1371/journal.pone.0220311. eCollection 2019. PLoS One. 2019. PMID: 31408468 Free PMC article.
References
-
- Ordonez-Moran P, Munoz A. Nuclear receptors: genomic and non-genomic effects converge. Cell Cycle. 2009;8:1675–1680. - PubMed
-
- Nassa G, Tarallo R, Ambrosino C, Bamundo A, Ferraro L, Paris O, Ravo M, Guzzi PH, Cannataro M, Baumann M, Nyman TA, Nola E, Weisz A. A large set of estrogen receptor beta-interacting proteins identified by tandem affinity purification in hormoneresponsive human breast cancer cell nuclei. Proteomics. in press . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

