Tetherin and its viral antagonists
- PMID: 21222046
- PMCID: PMC3087111
- DOI: 10.1007/s11481-010-9256-1
Tetherin and its viral antagonists
Abstract
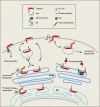
Restriction factors comprise an important layer of host defense to fight against viral infection. Some restriction factors are constitutively expressed whereas the majority is induced by interferon to elicit innate immunity. In addition to a number of well-characterized interferon-inducible antiviral factors such as RNaseL/OAS, ISG15, Mx, PKR, and ADAR, tetherin (BST-2/CD317/HM1.24) was recently discovered to block the release of enveloped viruses from the cell surface, which is regarded as a novel antiviral mechanism induced by interferon. Here, we briefly review the history of tetherin discovery, discuss how tetherin blocks virus production, and highlight the viral countermeasures to evade tetherin restriction.
Conflict of interest statement
There is no conflict of interest for any of the authors.
Figures

Similar articles
-
BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor.Mol Immunol. 2013 Jun;54(2):132-9. doi: 10.1016/j.molimm.2012.11.008. Epub 2012 Dec 27. Mol Immunol. 2013. PMID: 23274150 Review.
-
Limiting Respiratory Viral Infection by Targeting Antiviral and Immunological Functions of BST-2/Tetherin: Knowledge and Gaps.Bioessays. 2018 Oct;40(10):e1800086. doi: 10.1002/bies.201800086. Epub 2018 Aug 16. Bioessays. 2018. PMID: 30113067 Free PMC article. Review.
-
Influenza virus partially counteracts restriction imposed by tetherin/BST-2.J Biol Chem. 2012 Jun 22;287(26):22015-29. doi: 10.1074/jbc.M111.319996. Epub 2012 Apr 6. J Biol Chem. 2012. PMID: 22493439 Free PMC article.
-
Antiviral inhibition of enveloped virus release by tetherin/BST-2: action and counteraction.Viruses. 2011 May;3(5):520-40. doi: 10.3390/v3050520. Epub 2011 May 6. Viruses. 2011. PMID: 21994744 Free PMC article. Review.
-
HIV-1 Vpu Antagonizes CD317/Tetherin by Adaptor Protein-1-Mediated Exclusion from Virus Assembly Sites.J Virol. 2016 Jul 11;90(15):6709-6723. doi: 10.1128/JVI.00504-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27170757 Free PMC article.
Cited by
-
MT1-MMP Inhibits the Activity of Bst-2 via Their Cytoplasmic Domains Dependent Interaction.Int J Mol Sci. 2016 May 26;17(6):818. doi: 10.3390/ijms17060818. Int J Mol Sci. 2016. PMID: 27240342 Free PMC article.
-
HIV blocks Type I IFN signaling through disruption of STAT1 phosphorylation.Innate Immun. 2018 Nov;24(8):490-500. doi: 10.1177/1753425918803674. Epub 2018 Oct 3. Innate Immun. 2018. PMID: 30282499 Free PMC article.
-
Microbial Biofilms: Human T-cell Leukemia Virus Type 1 First in Line for Viral Biofilm but Far Behind Bacterial Biofilms.Front Microbiol. 2020 Sep 15;11:2041. doi: 10.3389/fmicb.2020.02041. eCollection 2020. Front Microbiol. 2020. PMID: 33042035 Free PMC article. Review.
-
Interferon-induced tetherin restricts vesicular stomatitis virus release in neurons.DNA Cell Biol. 2011 Dec;30(12):965-74. doi: 10.1089/dna.2011.1384. Epub 2011 Sep 15. DNA Cell Biol. 2011. PMID: 21919738 Free PMC article.
-
Host Cell Restriction Factors of Paramyxoviruses and Pneumoviruses.Viruses. 2020 Dec 2;12(12):1381. doi: 10.3390/v12121381. Viruses. 2020. PMID: 33276587 Free PMC article. Review.
References
-
- Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

