Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus
- PMID: 21220524
- PMCID: PMC3067146
- DOI: 10.1128/AAC.01213-10
Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus
Abstract
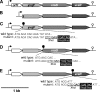
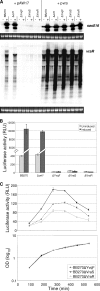
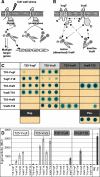
The exposure of Staphylococcus aureus to a broad range of cell wall-damaging agents triggers the induction of a cell wall stress stimulon (CWSS) controlled by the VraSR two-component system. The vraSR genes form part of the four-cistron autoregulatory operon orf1-yvqF-vraS-vraR. The markerless inactivation of each of the genes within this operon revealed that orf1 played no observable role in CWSS induction and had no influence on resistance phenotypes for any of the cell envelope stress-inducing agents tested. The remaining three genes were all essential for the induction of the CWSS, and mutants showed various degrees of increased susceptibility to cell wall-active antibiotics. Therefore, the role of YvqF in S. aureus appears to be opposite that in other Gram-positive bacteria, where YvqF homologs have all been shown to inhibit signal transduction. This role, as an activator rather than repressor of signal transduction, corresponds well with resistance phenotypes of ΔYvqF mutants, which were similar to those of ΔVraR mutants in which CWSS induction also was completely abolished. Resistance profiles of ΔVraS mutants differed phenotypically from those of ΔYvqF and ΔVraR mutants on many non-ß-lactam antibiotics. ΔVraS mutants still became more susceptible than wild-type strains at low antibiotic concentrations, but they retained larger subpopulations that were able to grow on higher antibiotic concentrations than ΔYvqF and ΔVraR mutants. Subpopulations of ΔVraS mutants could grow on even higher glycopeptide concentrations than wild-type strains. The expression of a highly sensitive CWSS-luciferase reporter gene fusion was up to 2.6-fold higher in a ΔVraS than a ΔVraR mutant, which could be linked to differences in their respective antibiotic resistance phenotypes. Bacterial two-hybrid analysis indicated that the integral membrane protein YvqF interacted directly with VraS but not VraR, suggesting that it plays an essential role in sensing the as-yet unknown trigger of CWSS induction.
Figures



Similar articles
-
Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics.BMC Microbiol. 2011 Jan 20;11:16. doi: 10.1186/1471-2180-11-16. BMC Microbiol. 2011. PMID: 21251258 Free PMC article.
-
VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus.Antimicrob Agents Chemother. 2013 Jan;57(1):83-95. doi: 10.1128/AAC.01651-12. Epub 2012 Oct 15. Antimicrob Agents Chemother. 2013. PMID: 23070169 Free PMC article.
-
Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR.J Antimicrob Chemother. 2010 Jan;65(1):37-45. doi: 10.1093/jac/dkp394. J Antimicrob Chemother. 2010. PMID: 19889788 Free PMC article.
-
Site-specific mutation of Staphylococcus aureus VraS reveals a crucial role for the VraR-VraS sensor in the emergence of glycopeptide resistance.Antimicrob Agents Chemother. 2011 Mar;55(3):1008-20. doi: 10.1128/AAC.00720-10. Epub 2010 Dec 20. Antimicrob Agents Chemother. 2011. PMID: 21173175 Free PMC article.
-
Chromatin immunoprecipitation identifies genes under direct VraSR regulation in Staphylococcus aureus.Can J Microbiol. 2012 Jun;58(6):703-8. doi: 10.1139/w2012-043. Epub 2012 May 9. Can J Microbiol. 2012. PMID: 22571705
Cited by
-
Investigation into the mechanism of action of the antimicrobial peptide epilancin 15X.Front Microbiol. 2023 Nov 2;14:1247222. doi: 10.3389/fmicb.2023.1247222. eCollection 2023. Front Microbiol. 2023. PMID: 38029153 Free PMC article.
-
Lantibiotic resistance.Microbiol Mol Biol Rev. 2015 Jun;79(2):171-91. doi: 10.1128/MMBR.00051-14. Microbiol Mol Biol Rev. 2015. PMID: 25787977 Free PMC article. Review.
-
Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300.PLoS Pathog. 2012 Feb;8(2):e1002505. doi: 10.1371/journal.ppat.1002505. Epub 2012 Feb 2. PLoS Pathog. 2012. PMID: 22319446 Free PMC article.
-
Long-Term Intrahost Evolution of Staphylococcus aureus Among Diabetic Patients With Foot Infections.Front Microbiol. 2021 Sep 6;12:741406. doi: 10.3389/fmicb.2021.741406. eCollection 2021. Front Microbiol. 2021. PMID: 34552578 Free PMC article.
-
A vancomycin resistance-associated WalK(S221P) mutation attenuates the virulence of vancomycin-intermediate Staphylococcus aureus.J Adv Res. 2022 Sep;40:167-178. doi: 10.1016/j.jare.2021.11.015. Epub 2021 Nov 26. J Adv Res. 2022. PMID: 36100324 Free PMC article.
References
-
- Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. - PubMed
-
- Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system: a mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. - PubMed
-
- Belcheva, A., V. Verma, and D. Golemi-Kotra. 2009. DNA-binding activity of the vancomycin resistance associated regulator protein VraR and the role of phosphorylation in transcriptional regulation of the vraSR operon. Biochemistry 48:5592-5601. - PubMed
-
- Berger-Bächi, B., and M. L. Kohler. 1983. A novel site on the chromosome of Staphylococcus aureus influencing the level of methicillin resistance: genetic mapping. FEMS Microbiol. Lett. 20:305-309.
-
- Blake, K. L., et al. 2009. The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol. Microbiol. 72:335-343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

