Reaction mechanism of single subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae: evidence for a ternary complex mechanism
- PMID: 21220430
- PMCID: PMC3059053
- DOI: 10.1074/jbc.M110.175547
Reaction mechanism of single subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae: evidence for a ternary complex mechanism
Abstract
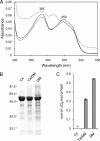
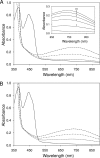
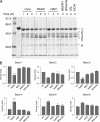
The flavoprotein rotenone-insensitive internal NADH-ubiquinone (UQ) oxidoreductase (Ndi1) is a member of the respiratory chain in Saccharomyces cerevisiae. We reported previously that bound UQ in Ndi1 plays a key role in preventing the generation of reactive oxygen species. Here, to elucidate this mechanism, we investigated biochemical properties of Ndi1 and its mutants in which highly conserved amino acid residues (presumably involved in NADH and/or UQ binding sites) were replaced. We found that wild-type Ndi1 formed a stable charge transfer (CT) complex (around 740 nm) with NADH, but not with NADPH, under anaerobic conditions. The intensity of the CT absorption band was significantly increased by the presence of bound UQ or externally added n-decylbenzoquinone. Interestingly, however, when Ndi1 was exposed to air, the CT band transiently reached the same maximum level regardless of the presence of UQ. This suggests that Ndi1 forms a ternary complex with NADH and UQ, but the role of UQ in withdrawing an electron can be substitutable with oxygen. Proteinase K digestion analysis showed that NADH (but not NADPH) binding induces conformational changes in Ndi1. The kinetic study of wild-type and mutant Ndi1 indicated that there is no overlap between NADH and UQ binding sites. Moreover, we found that the bound UQ can reversibly dissociate from Ndi1 and is thus replaceable with other quinones in the membrane. Taken together, unlike other NAD(P)H-UQ oxidoreductases, the Ndi1 reaction proceeds through a ternary complex (not a ping-pong) mechanism. The bound UQ keeps oxygen away from the reduced flavin.
Figures


Similar articles
-
Structural insight into the type-II mitochondrial NADH dehydrogenases.Nature. 2012 Nov 15;491(7424):478-82. doi: 10.1038/nature11541. Epub 2012 Oct 21. Nature. 2012. PMID: 23086143
-
New complexes containing the internal alternative NADH dehydrogenase (Ndi1) in mitochondria of Saccharomyces cerevisiae.Yeast. 2015 Oct;32(10):629-41. doi: 10.1002/yea.3086. Epub 2015 Aug 21. Yeast. 2015. PMID: 26173916
-
Roles of bound quinone in the single subunit NADH-quinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae.J Biol Chem. 2007 Mar 2;282(9):6012-20. doi: 10.1074/jbc.M610646200. Epub 2007 Jan 2. J Biol Chem. 2007. PMID: 17200125
-
Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex I.Biochim Biophys Acta. 2016 Jul;1857(7):872-83. doi: 10.1016/j.bbabio.2015.12.009. Epub 2015 Dec 22. Biochim Biophys Acta. 2016. PMID: 26721206 Free PMC article. Review.
-
Kinetics, control, and mechanism of ubiquinone reduction by the mammalian respiratory chain-linked NADH-ubiquinone reductase.J Bioenerg Biomembr. 1993 Aug;25(4):367-75. doi: 10.1007/BF00762462. J Bioenerg Biomembr. 1993. PMID: 8226718 Review.
Cited by
-
Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors.Sci Rep. 2018 Feb 5;8(1):2427. doi: 10.1038/s41598-018-20775-6. Sci Rep. 2018. PMID: 29402945 Free PMC article.
-
Structural and Functional insights into the catalytic mechanism of the Type II NADH:quinone oxidoreductase family.Sci Rep. 2017 Feb 9;7:42303. doi: 10.1038/srep42303. Sci Rep. 2017. PMID: 28181562 Free PMC article.
-
Expression of yeast NDI1 rescues a Drosophila complex I assembly defect.PLoS One. 2012;7(11):e50644. doi: 10.1371/journal.pone.0050644. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226344 Free PMC article.
-
Mycobacterium tuberculosis type II NADH-menaquinone oxidoreductase catalyzes electron transfer through a two-site ping-pong mechanism and has two quinone-binding sites.Biochemistry. 2014 Feb 25;53(7):1179-90. doi: 10.1021/bi4013897. Epub 2014 Feb 11. Biochemistry. 2014. PMID: 24447297 Free PMC article.
-
The oligomeric state of the Caldivirga maquilingensis type III sulfide:Quinone Oxidoreductase is required for membrane binding.Biochim Biophys Acta Bioenerg. 2020 Feb 1;1861(2):148132. doi: 10.1016/j.bbabio.2019.148132. Epub 2019 Dec 6. Biochim Biophys Acta Bioenerg. 2020. PMID: 31816290 Free PMC article.
References
-
- Yagi T., Di Bernardo S., Nakamaru-Ogiso E., Kao M. C., Seo B. B., Matsuno-Yagi A. (2004) in Respiration in Archaea and Bacteria (Zannori D. ed) pp. 15–40, Kluwer Academic Publishers, Dordrecht, The Netherlands
-
- Moller I. M. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591 - PubMed
-
- Yagi T., Seo B. B., Nakamaru-Ogiso E., Marella M., Barber-Singh J., Yamashita T., Kao M. C., Matsuno-Yagi A. (2006) Rejuvenation Res. 9, 191–197 - PubMed
-
- Yamashita T., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2007) J. Biol. Chem. 282, 6012–6020 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

