A hybrid BPSO-CGA approach for gene selection and classification of microarray data
- PMID: 21210743
- PMCID: PMC3244808
- DOI: 10.1089/cmb.2010.0064
A hybrid BPSO-CGA approach for gene selection and classification of microarray data
Abstract
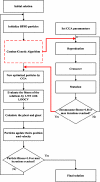
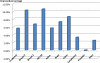
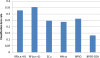
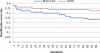
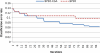
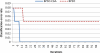
Microarray analysis promises to detect variations in gene expressions, and changes in the transcription rates of an entire genome in vivo. Microarray gene expression profiles indicate the relative abundance of mRNA corresponding to the genes. The selection of relevant genes from microarray data poses a formidable challenge to researchers due to the high-dimensionality of features, multiclass categories being involved, and the usually small sample size. A classification process is often employed which decreases the dimensionality of the microarray data. In order to correctly analyze microarray data, the goal is to find an optimal subset of features (genes) which adequately represents the original set of features. A hybrid method of binary particle swarm optimization (BPSO) and a combat genetic algorithm (CGA) is to perform the microarray data selection. The K-nearest neighbor (K-NN) method with leave-one-out cross-validation (LOOCV) served as a classifier. The proposed BPSO-CGA approach is compared to ten microarray data sets from the literature. The experimental results indicate that the proposed method not only effectively reduce the number of genes expression level, but also achieves a low classification error rate.
Figures



Similar articles
-
Chaotic genetic algorithm for gene selection and classification problems.OMICS. 2009 Oct;13(5):407-20. doi: 10.1089/omi.2009.0007. OMICS. 2009. PMID: 19594377
-
Tabu search and binary particle swarm optimization for feature selection using microarray data.J Comput Biol. 2009 Dec;16(12):1689-703. doi: 10.1089/cmb.2007.0211. J Comput Biol. 2009. PMID: 20047491
-
Correlation-based gene selection and classification using Taguchi-BPSO.Methods Inf Med. 2010;49(3):254-68. doi: 10.3414/ME09-01-0010. Epub 2010 Feb 5. Methods Inf Med. 2010. PMID: 20135079
-
Filter versus wrapper gene selection approaches in DNA microarray domains.Artif Intell Med. 2004 Jun;31(2):91-103. doi: 10.1016/j.artmed.2004.01.007. Artif Intell Med. 2004. PMID: 15219288 Review.
-
Gene set enrichment analysis: performance evaluation and usage guidelines.Brief Bioinform. 2012 May;13(3):281-91. doi: 10.1093/bib/bbr049. Epub 2011 Sep 7. Brief Bioinform. 2012. PMID: 21900207 Free PMC article. Review.
Cited by
-
DepthTools: an R package for a robust analysis of gene expression data.BMC Bioinformatics. 2013 Jul 25;14:237. doi: 10.1186/1471-2105-14-237. BMC Bioinformatics. 2013. PMID: 23885712 Free PMC article.
-
Feature Selection and Feature Stability Measurement Method for High-Dimensional Small Sample Data Based on Big Data Technology.Comput Intell Neurosci. 2021 Sep 23;2021:3597051. doi: 10.1155/2021/3597051. eCollection 2021. Comput Intell Neurosci. 2021. PMID: 34603430 Free PMC article.
-
Antimicrobial Resistance and Genomic Characterization of Six New Sequence Types in Multidrug-Resistant Pseudomonas aeruginosa Clinical Isolates from Pakistan.Antibiotics (Basel). 2021 Nov 12;10(11):1386. doi: 10.3390/antibiotics10111386. Antibiotics (Basel). 2021. PMID: 34827324 Free PMC article.
-
Supervised Methods for Biomarker Detection from Microarray Experiments.Methods Mol Biol. 2022;2401:101-120. doi: 10.1007/978-1-0716-1839-4_8. Methods Mol Biol. 2022. PMID: 34902125
-
A graph-based gene selection method for medical diagnosis problems using a many-objective PSO algorithm.BMC Med Inform Decis Mak. 2021 Nov 27;21(1):333. doi: 10.1186/s12911-021-01696-3. BMC Med Inform Decis Mak. 2021. PMID: 34838034 Free PMC article.
References
-
- AltIncay H. Ensembling evidential k-nearest neighbor classifiers through multi-modal perturbation. Applied Soft Computing. 2007;7:1072–1083.
-
- Breiman L. Classification and Regression Trees. Chapman & Hall/CRC; Boca Raton, FL: 1984.
-
- Buturovic L.J. PCP: a program for supervised classification of gene expression profiles. Bioinformatics. 2006;22:245. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

