Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces
- PMID: 21205160
- PMCID: PMC3079286
- DOI: 10.1111/j.1567-1364.2010.00712.x
Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces
Abstract
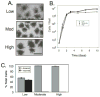
Saccharomyces cerevisiae grown on plastic surfaces formed organized structures, termed minicolonies, that consisted of a core of round (yeast-like) cells surrounded by chains of filamentous cells (pseudohyphae). Minicolonies had a much higher affinity for plastic than unstructured yeast communities growing on the same surface. Pseudohyphae at the surface of these colonies developed further into chains of asci. These structures suggest that pseudohyphal differentiation and sporulation are sequential processes in minicolonies. Consistent with this idea, minicolonies grown under conditions that stimulated pseudohyphal differentiation contained higher frequencies of asci. Furthermore, a flo11Δ mutant, which fails to form pseudohyphae, yielded normal sporulation in cultures, but was defective for minicolony sporulation. When minicolonies were dispersed in water and cells were then allowed to settle on the plastic surface, these cells sporulated very efficiently. Taken together, our results suggest that sporulation in minicolonies is stimulated by pseudohyphal differentiation because these pseudohyphae are dispersed from the core of the colony.
© 2011 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures



Similar articles
-
Role of distinct dimorphic transitions in territory colonizing and formation of yeast colony architecture.Environ Microbiol. 2010 Jan;12(1):264-77. doi: 10.1111/j.1462-2920.2009.02067.x. Epub 2009 Oct 2. Environ Microbiol. 2010. PMID: 19799621
-
Stable Pseudohyphal Growth in Budding Yeast Induced by Synergism between Septin Defects and Altered MAP-kinase Signaling.PLoS Genet. 2015 Dec 7;11(12):e1005684. doi: 10.1371/journal.pgen.1005684. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26640955 Free PMC article.
-
Messengers for morphogenesis: inositol polyphosphate signaling and yeast pseudohyphal growth.Curr Genet. 2019 Feb;65(1):119-125. doi: 10.1007/s00294-018-0874-0. Epub 2018 Aug 12. Curr Genet. 2019. PMID: 30101372 Review.
-
Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae.J Cell Sci. 2005 Jul 1;118(Pt 13):2935-47. doi: 10.1242/jcs.02414. J Cell Sci. 2005. PMID: 15976451
-
Cross-talk between autophagy and sporulation in Saccharomyces cerevisiae.Yeast. 2021 Jul;38(7):401-413. doi: 10.1002/yea.3556. Epub 2021 Mar 15. Yeast. 2021. PMID: 33608896 Review.
Cited by
-
Aspects of Multicellularity in Saccharomyces cerevisiae Yeast: A Review of Evolutionary and Physiological Mechanisms.Genes (Basel). 2020 Jun 24;11(6):690. doi: 10.3390/genes11060690. Genes (Basel). 2020. PMID: 32599749 Free PMC article. Review.
-
Cell signals, cell contacts, and the organization of yeast communities.Eukaryot Cell. 2011 Apr;10(4):466-73. doi: 10.1128/EC.00313-10. Epub 2011 Feb 4. Eukaryot Cell. 2011. PMID: 21296916 Free PMC article. Review.
-
An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae.Genetics. 2014 Mar;196(3):853-65. doi: 10.1534/genetics.113.160291. Epub 2013 Dec 27. Genetics. 2014. PMID: 24374355 Free PMC article.
-
Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation.Microb Cell. 2016 Aug;3(8):302-328. doi: 10.15698/mic2016.08.516. Microb Cell. 2016. PMID: 27917388 Free PMC article.
-
Biofilm localization in the vertical wall of shaking 96-well plates.Scientifica (Cairo). 2014;2014:231083. doi: 10.1155/2014/231083. Epub 2014 Apr 13. Scientifica (Cairo). 2014. PMID: 24834360 Free PMC article.
References
-
- Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

