The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions
- PMID: 21194420
- PMCID: PMC3022720
- DOI: 10.1186/1471-2121-11-101
The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions
Abstract
Background: Chromatin insulators or boundary elements are a class of functional elements in the eukaryotic genome. They regulate gene transcription by interfering with promoter-enhancer communication. The Cp190 protein of Drosophila melanogaster is essential to the function of at least three-types of chromatin insulator complexes organized by Su(Hw), CTCF and BEAF32.
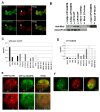
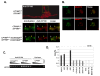
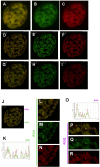
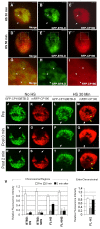
Results: We mapped functional regions of Cp190 in vivo and identified three domains that are essential for the insulator function and for the viability of flies: the BTB/POZ domain, an aspartic acid-rich (D-rich) region and a C-terminal glutamic acid-rich (E-rich) region. Other domains including the centrosomal targeting domain and the zinc fingers are dispensable. The N-terminal CP190BTB-D fragment containing the BTB/POZ domain and the D-rich region is sufficient to mediate association with all three types of insulator complexes. The fragment however is not sufficient for insulator activity or viability. The Cp190 and CP190BTB-D are regulated differently in cells treated with heat-shock. The Cp190 dissociated from chromosomes during heat-shock, indicating that dissociation of Cp190 with chromosomes can be regulated. In contrast, the CP190BTB-D fragment didn't dissociate from chromosomes in the same heat-shocked condition, suggesting that the deleted C-terminal regions have a role in regulating the dissociation of Cp190 with chromosomes.
Conclusions: The N-terminal fragment of Cp190 containing the BTB/POZ domain and the D-rich region mediates association of Cp190 with all three types of insulator complexes and that the E-rich region of Cp190 is required for dissociation of Cp190 from chromosomes during heat-shock. The heat-shock-induced dissociation is strong evidence indicating that dissociation of the essential insulator protein Cp190 from chromosomes is regulated. Our results provide a mechanism through which activities of an insulator can be modulated by internal and external cues.
Figures

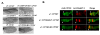

Similar articles
-
Interactions between BTB domain of CP190 and two adjacent regions in Su(Hw) are required for the insulator complex formation.Chromosoma. 2018 Mar;127(1):59-71. doi: 10.1007/s00412-017-0645-6. Epub 2017 Sep 22. Chromosoma. 2018. PMID: 28939920
-
Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome.PLoS Genet. 2014 Aug 28;10(8):e1004544. doi: 10.1371/journal.pgen.1004544. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25165871 Free PMC article.
-
Mechanisms of CP190 Interaction with Architectural Proteins in Drosophila Melanogaster.Int J Mol Sci. 2021 Nov 17;22(22):12400. doi: 10.3390/ijms222212400. Int J Mol Sci. 2021. PMID: 34830280 Free PMC article.
-
Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review.
-
Chromatin insulators: regulatory mechanisms and epigenetic inheritance.Mol Cell. 2008 Oct 10;32(1):1-9. doi: 10.1016/j.molcel.2008.08.017. Mol Cell. 2008. PMID: 18851828 Free PMC article. Review.
Cited by
-
Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains.Genome Res. 2012 Nov;22(11):2176-87. doi: 10.1101/gr.136788.111. Epub 2012 Jun 21. Genome Res. 2012. PMID: 22722341 Free PMC article.
-
Distinct Roles of Chromatin Insulator Proteins in Control of the Drosophila Bithorax Complex.Genetics. 2016 Feb;202(2):601-17. doi: 10.1534/genetics.115.179309. Epub 2015 Dec 29. Genetics. 2016. PMID: 26715665 Free PMC article.
-
Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin.Genome Res. 2015 Jan;25(1):89-99. doi: 10.1101/gr.174169.114. Epub 2014 Oct 23. Genome Res. 2015. PMID: 25342723 Free PMC article.
-
Interactions between BTB domain of CP190 and two adjacent regions in Su(Hw) are required for the insulator complex formation.Chromosoma. 2018 Mar;127(1):59-71. doi: 10.1007/s00412-017-0645-6. Epub 2017 Sep 22. Chromosoma. 2018. PMID: 28939920
-
The Functions and Mechanisms of Action of Insulators in the Genomes of Higher Eukaryotes.Acta Naturae. 2020 Oct-Dec;12(4):15-33. doi: 10.32607/actanaturae.11144. Acta Naturae. 2020. PMID: 33456975 Free PMC article.
References
-
- Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113(3):735–747. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

