Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt
- PMID: 21191028
- PMCID: PMC3067963
- DOI: 10.1128/JVI.02175-10
Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt
Abstract
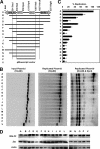
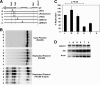
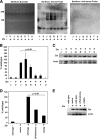
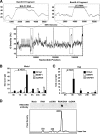
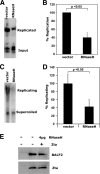
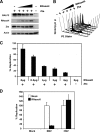
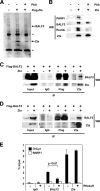
The genetic elements of herpesvirus origins of lytic replication have been characterized in detail; however, much remains to be elucidated concerning their functional role in replication initiation. In the case of the Epstein-Barr virus (EBV), we have found that in addition to the two well-defined critical elements required for lytic replication (the upstream and downstream essential elements, UEE and DEE), the origin of lytic replication (OriLyt) also requires the presence of a GC-rich RNA in cis. The BHLF1 transcript is similar to the essential K5 transcript identified at the Kaposi's sarcoma-associated herpesvirus OriLyt. We have found that truncation of the BHLF1 transcript or deletion of the TATA box, but not the putative ATG initiation codon, reduce OriLyt function to background levels. By using an antibody specific for RNA-DNA hybrid molecules, we found the BHLF1 RNA stably annealed to its DNA template during the early steps of lytic reactivation. Furthermore, expression of human RNase H1, which degrades RNA in RNA-DNA hybrids, drastically reduces OriLyt-dependent DNA replication as well as recruitment of the viral single-stranded DNA binding protein BALF2 to OriLyt. These studies suggest that a GC-rich OriLyt transcript is an important component of gammaherpesvirus lytic origins and is required for initial strand separation and loading of core replication proteins.
Figures

Similar articles
-
The SWI/SNF Chromatin Regulator BRG1 Modulates the Transcriptional Regulatory Activity of the Epstein-Barr Virus DNA Polymerase Processivity Factor BMRF1.J Virol. 2017 Apr 13;91(9):e02114-16. doi: 10.1128/JVI.02114-16. Print 2017 May 1. J Virol. 2017. PMID: 28228591 Free PMC article.
-
Evidence for DNA hairpin recognition by Zta at the Epstein-Barr virus origin of lytic replication.J Virol. 2010 Jul;84(14):7073-82. doi: 10.1128/JVI.02666-09. Epub 2010 May 5. J Virol. 2010. PMID: 20444899 Free PMC article.
-
Complexities associated with expression of Epstein-Barr virus (EBV) lytic origins of DNA replication.Nucleic Acids Res. 2007;35(10):3391-406. doi: 10.1093/nar/gkm170. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478522 Free PMC article.
-
Nuts and bolts of human cytomegalovirus lytic DNA replication.Curr Top Microbiol Immunol. 2008;325:153-66. doi: 10.1007/978-3-540-77349-8_9. Curr Top Microbiol Immunol. 2008. PMID: 18637505 Review.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
Cited by
-
Regulation of R-Loops in DNA Tumor Viruses.Pathogens. 2024 Oct 2;13(10):863. doi: 10.3390/pathogens13100863. Pathogens. 2024. PMID: 39452734 Free PMC article. Review.
-
Long noncoding RNAs involvement in Epstein-Barr virus infection and tumorigenesis.Virol J. 2020 Apr 9;17(1):51. doi: 10.1186/s12985-020-01308-y. Virol J. 2020. PMID: 32272952 Free PMC article. Review.
-
The SWI/SNF Chromatin Regulator BRG1 Modulates the Transcriptional Regulatory Activity of the Epstein-Barr Virus DNA Polymerase Processivity Factor BMRF1.J Virol. 2017 Apr 13;91(9):e02114-16. doi: 10.1128/JVI.02114-16. Print 2017 May 1. J Virol. 2017. PMID: 28228591 Free PMC article.
-
Genome-wide analysis of Epstein-Barr virus Rta DNA binding.J Virol. 2012 May;86(9):5151-64. doi: 10.1128/JVI.06760-11. Epub 2012 Feb 29. J Virol. 2012. PMID: 22379087 Free PMC article.
-
Clinical significance of molecular subtypes of gastrointestinal tract adenocarcinoma.World J Gastrointest Oncol. 2022 Mar 15;14(3):628-645. doi: 10.4251/wjgo.v14.i3.628. World J Gastrointest Oncol. 2022. PMID: 35321271 Free PMC article. Review.
References
-
- Billaud, G., D. Thouvenot, and F. Morfin. 2009. Drug targets in herpes simplex and Epstein Barr virus infections. Infect. Disord. Drug Targets 9:117-125. - PubMed
-
- Boguslawski, S. J., et al. 1986. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89:123-130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

