EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5
- PMID: 21177815
- PMCID: PMC3067769
- DOI: 10.1128/JVI.02279-10
EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5
Abstract
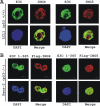
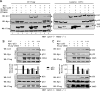
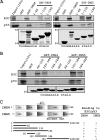
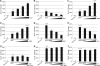
Epstein-Barr virus (EBV)-encoded EBNA3C is one of the latent proteins essential for the efficient transformation of human primary B lymphocytes into continuously proliferating lymphoblastoid cell lines (LCLs) in vitro through manipulation of a number of major cellular pathways. Although it does not have direct DNA-binding activity, EBNA3C plays a central role in the transcriptional modulation of a wide range of both viral and cellular genes during latent infection. Recently, we showed that EBNA3C can directly bind to the tumor suppressor protein p53 and repress its functions, in part by blocking its transcriptional activity as well as facilitating its degradation through stabilization of its negative regulator, Mdm2. In this study, we further showed that EBNA3C can negatively regulate p53-mediated functions by interacting with its regulatory proteins, the inhibitor of growth family proteins ING4 and ING5, shown to be frequently deregulated in different cancers. Functional mapping revealed that both ING4 and ING5 bound to N-terminal domain residues 129 to 200 of EBNA3C, which was previously demonstrated to associate with p53 and is also essential for LCL growth. In addition, we showed that a conserved domain of either ING4 or ING5 bound to both p53 and EBNA3C in a competitive manner, suggesting a potential role for EBNA3C whereby the ING4 or -5/p53 pathway is modulated in EBV-infected cells. Subsequently, we demonstrated that EBNA3C significantly suppresses both the ING4- and ING5-mediated regulation of p53 transcriptional activity in a dose-dependent manner. A colony formation assay as well as an apoptosis assay showed that EBNA3C nullified the negative regulatory effects on cell proliferation induced by coupled expression of p53 in the presence of either ING4 or ING5 in Saos-2 (p53(-/-)) cells. This report demonstrates a possible role for the candidate tumor suppressor ING genes in the biology of EBV-associated cancers.
Figures


Similar articles
-
Epstein-Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis.PLoS Pathog. 2011 Dec;7(12):e1002418. doi: 10.1371/journal.ppat.1002418. Epub 2011 Dec 8. PLoS Pathog. 2011. Retraction in: PLoS Pathog. 2021 Apr 30;17(4):e1009556. doi: 10.1371/journal.ppat.1009556. PMID: 22174681 Free PMC article. Retracted.
-
Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2.J Virol. 2009 May;83(9):4652-69. doi: 10.1128/JVI.02408-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244339 Free PMC article.
-
E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells.PLoS Pathog. 2012;8(3):e1002573. doi: 10.1371/journal.ppat.1002573. Epub 2012 Mar 15. PLoS Pathog. 2012. PMID: 22438805 Free PMC article.
-
The EBNA3 Family: Two Oncoproteins and a Tumour Suppressor that Are Central to the Biology of EBV in B Cells.Curr Top Microbiol Immunol. 2015;391:61-117. doi: 10.1007/978-3-319-22834-1_3. Curr Top Microbiol Immunol. 2015. PMID: 26428372 Review.
-
Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus.Mol Cell Biochem. 2009 Sep;329(1-2):131-9. doi: 10.1007/s11010-009-0123-4. Epub 2009 May 3. Mol Cell Biochem. 2009. PMID: 19412732 Free PMC article. Review.
Cited by
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
-
EBNA3C facilitates RASSF1A downregulation through ubiquitin-mediated degradation and promoter hypermethylation to drive B-cell proliferation.PLoS Pathog. 2019 Jan 7;15(1):e1007514. doi: 10.1371/journal.ppat.1007514. eCollection 2019 Jan. PLoS Pathog. 2019. PMID: 30615685 Free PMC article.
-
Epstein-Barr Virus (EBV)-Related Lymphoproliferative Disorders in Ataxia Telangiectasia: Does ATM Regulate EBV Life Cycle?Front Immunol. 2019 Jan 4;9:3060. doi: 10.3389/fimmu.2018.03060. eCollection 2018. Front Immunol. 2019. PMID: 30662441 Free PMC article. Review.
-
Transcriptional and epigenetic modulation of autophagy promotes EBV oncoprotein EBNA3C induced B-cell survival.Cell Death Dis. 2018 May 22;9(6):605. doi: 10.1038/s41419-018-0668-9. Cell Death Dis. 2018. PMID: 29789559 Free PMC article.
-
ING5 Inhibits Migration and Invasion of Esophageal Cancer Cells by Downregulating the IL-6/CXCL12 Signaling Pathway.Technol Cancer Res Treat. 2021 Jan-Dec;20:15330338211039940. doi: 10.1177/15330338211039940. Technol Cancer Res Treat. 2021. PMID: 34520285 Free PMC article.
References
-
- Doyon, Y., et al. 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21:51-64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

