Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria
- PMID: 21173275
- PMCID: PMC3017135
- DOI: 10.1073/pnas.1014918108
Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria
Abstract
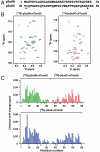
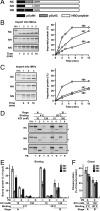
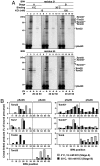
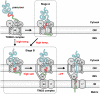
Mitochondria import most of their resident proteins from the cytosol, and the import receptor Tom20 of the outer-membrane translocator TOM40 complex plays an essential role in specificity of mitochondrial protein import. Here we analyzed the effects of Tom20 binding on NMR spectra of a long mitochondrial presequence and found that it contains two distinct Tom20-binding elements. In vitro import and cross-linking experiments revealed that, although the N-terminal Tom20-binding element is essential for targeting to mitochondria, the C-terminal element increases efficiency of protein import in the step prior to translocation across the inner membrane. Therefore Tom20 has a dual role in protein import into mitochondria: recognition of the targeting signal in the presequence and tethering the presequence to the TOM40 complex to increase import efficiency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein.J Biol Chem. 1999 Jun 4;274(23):16522-30. doi: 10.1074/jbc.274.23.16522. J Biol Chem. 1999. PMID: 10347216
-
Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import.J Biol Chem. 2008 Feb 15;283(7):3799-807. doi: 10.1074/jbc.M708339200. Epub 2007 Dec 6. J Biol Chem. 2008. PMID: 18063580
-
Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence.J Biol Chem. 1997 Jul 25;272(30):18725-31. doi: 10.1074/jbc.272.30.18725. J Biol Chem. 1997. PMID: 9228044
-
Functions of outer membrane receptors in mitochondrial protein import.Biochim Biophys Acta. 2002 Sep 2;1592(1):3-14. doi: 10.1016/s0167-4889(02)00259-8. Biochim Biophys Acta. 2002. PMID: 12191763 Review.
-
How to get to the other side of the mitochondrial inner membrane - the protein import motor.Biol Chem. 2020 May 26;401(6-7):723-736. doi: 10.1515/hsz-2020-0106. Biol Chem. 2020. PMID: 32142474 Review.
Cited by
-
MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites.Mol Cell Proteomics. 2015 Apr;14(4):1113-26. doi: 10.1074/mcp.M114.043083. Epub 2015 Feb 10. Mol Cell Proteomics. 2015. PMID: 25670805 Free PMC article.
-
FgRab5 and FgRab7 are essential for endosomes biogenesis and non-redundantly recruit the retromer complex to the endosomes in Fusarium graminearum.Stress Biol. 2021 Dec 6;1(1):17. doi: 10.1007/s44154-021-00020-3. Stress Biol. 2021. PMID: 37676350 Free PMC article.
-
Mitochondrial abnormalities in temporal lobe of autistic brain.Neurobiol Dis. 2013 Jun;54:349-61. doi: 10.1016/j.nbd.2013.01.006. Epub 2013 Jan 17. Neurobiol Dis. 2013. PMID: 23333625 Free PMC article.
-
Identification of H3N2 NA and PB1-F2 genetic variants and their association with disease symptoms during the 2014-15 influenza season.Virus Evol. 2021 Jun 4;7(1):veab047. doi: 10.1093/ve/veab047. eCollection 2021 Jan. Virus Evol. 2021. PMID: 34131512 Free PMC article.
-
Biogenesis of Mitochondrial Metabolite Carriers.Biomolecules. 2020 Jul 7;10(7):1008. doi: 10.3390/biom10071008. Biomolecules. 2020. PMID: 32645990 Free PMC article. Review.
References
-
- Schatz G, Dobberstein N. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. - PubMed
-
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. - PubMed
-
- Koehler CM. New developments in mitochondrial assembly. Annu Rev Cell Dev Biol. 2004;20:309–335. - PubMed
-
- Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

