Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation
- PMID: 21150308
- PMCID: PMC3092684
- DOI: 10.4161/epi.6.3.14282
Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation
Abstract
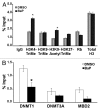
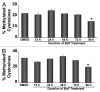
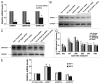
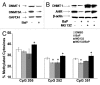
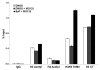
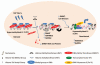
Benzo(a)pyrene (BaP), is an environmental pollutant present in tobacco smoke and a byproduct of fossil fuel combustion which likely contributes to the tumorigenic processes in human cancers including lung and esophageal. Long Interspersed Nuclear Element-1 (LINE-1) or L1 is a mobile element within the mammalian genome that propagates via a "copy-and-paste" mechanism using reverse transcriptase and RNA intermediates. L1 is strongly expressed during early embryogenesis and then silenced as cells initiate differentiation programming. Although the complex transcriptional control mechanisms of L1 are not well understood, L1 reactivation has been described in several human cancers and following exposure of mouse or human cells to BaP. In this study we investigated the molecular mechanisms and epigenetic events that regulate L1 reactivation following BaP exposure. We show that challenge of HeLa cells with BaP induces early enrichment of the transcriptionally-active chromatin markers histone H3 trimethylated at lysine 4 (H3K4Me3) and histone H3 acetylated at lysine 9 (H3K9Ac), and reduces association of DNA methyltransferase-1 (DNMT1) with the L1 promoter. These changes are followed by proteasome-dependent decreases in cellular DNMT1 expression and sustained reduction of cytosine methylation within the L1 promoter CpG island. Pharmacological inhibition of the proteasome signaling pathway with the inhibitor MG132 blocks degradation of DNMT1 and alters BaP-mediated histone epigenetic modifications. We conclude that genetic reactivation of L1 by BaP involves an ordered cascade of epigenetic events that begin with nucleosomal histone modifications and is completed with alterations in DNMT1 recruitment to the L1 promoter and reduced DNA methylation of CpG islands.
Figures

Similar articles
-
Influence of Benzo(a)pyrene on Different Epigenetic Processes.Int J Mol Sci. 2021 Dec 15;22(24):13453. doi: 10.3390/ijms222413453. Int J Mol Sci. 2021. PMID: 34948252 Free PMC article. Review.
-
H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1708-13. doi: 10.1073/pnas.1213266110. Epub 2013 Jan 9. Proc Natl Acad Sci U S A. 2013. PMID: 23302691 Free PMC article.
-
Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter.Mol Cell Biol. 2008 May;28(10):3219-35. doi: 10.1128/MCB.01516-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332107 Free PMC article.
-
Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen.Cancer Res. 2006 Mar 1;66(5):2616-20. doi: 10.1158/0008-5472.CAN-05-3478. Cancer Res. 2006. PMID: 16510580
-
Regulation of maintenance DNA methylation via histone ubiquitylation.J Biochem. 2016 Jan;159(1):9-15. doi: 10.1093/jb/mvv113. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590302 Free PMC article. Review.
Cited by
-
Sequence-specific biosensors report drug-induced changes in epigenetic silencing in living cells.DNA Cell Biol. 2012 Oct;31 Suppl 1(Suppl 1):S2-10. doi: 10.1089/dna.2011.1537. Epub 2012 Feb 7. DNA Cell Biol. 2012. PMID: 22313050 Free PMC article.
-
Modified CDKN2B (p15) and CDKN2A (p16) DNA methylation profiles in urban pesticide applicators.Environ Sci Pollut Res Int. 2019 May;26(15):15124-15135. doi: 10.1007/s11356-019-04658-5. Epub 2019 Mar 28. Environ Sci Pollut Res Int. 2019. PMID: 30924039
-
Long Interspersed Nuclear Element-1 Analytes in Extracellular Vesicles as Tools for Molecular Diagnostics of Non-Small Cell Lung Cancer.Int J Mol Sci. 2024 Jan 18;25(2):1169. doi: 10.3390/ijms25021169. Int J Mol Sci. 2024. PMID: 38256242 Free PMC article.
-
Environmental epigenetics and its implication on disease risk and health outcomes.ILAR J. 2012;53(3-4):289-305. doi: 10.1093/ilar.53.3-4.289. ILAR J. 2012. PMID: 23744968 Free PMC article. Review.
-
Epigenetic control of embryonic renal cell differentiation by L1 retrotransposon.Birth Defects Res A Clin Mol Teratol. 2011 Aug;91(8):693-702. doi: 10.1002/bdra.20786. Epub 2011 Mar 7. Birth Defects Res A Clin Mol Teratol. 2011. PMID: 21384534 Free PMC article.
References
-
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. - PubMed
-
- Feng Q, Moran JV, Kazazian H, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. - PubMed
-
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
