Dynamic conformations of the CD38-mediated NAD cyclization captured in a single crystal
- PMID: 21134381
- PMCID: PMC3019291
- DOI: 10.1016/j.jmb.2010.11.044
Dynamic conformations of the CD38-mediated NAD cyclization captured in a single crystal
Abstract
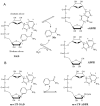
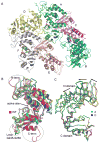
The extracellular domain of human CD38 is a multifunctional enzyme involved in the metabolism of two Ca(2+) messengers: cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate. When NAD is used as substrate, CD38 predominantly hydrolyzes it to ADP-ribose, with a trace amount of cyclic ADP-ribose produced through cyclization of the substrate. However, mutation of a key residue at the active site, E146, inhibits the hydrolysis activity of CD38 but greatly increases its cyclization activity. To understand the role of the residue E146 in the catalytic process, we determined the crystal structure of the E146A mutant protein with a substrate analogue, arabinosyl-2'-fluoro-deoxy-nicotinamide adenine dinucleotide. The structure captured the enzymatic reaction intermediates in six different conformations in a crystallographic asymmetric unit. The structural results indicate a folding-back process for the adenine ring of the substrate and provide the first multiple snapshots of the process. Our approach of utilizing multiple molecules in the crystallographic asymmetric unit should be generally applicable for capturing the dynamic nature of enzymatic catalysis.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
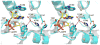

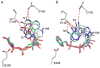
Similar articles
-
Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization.J Biol Chem. 2012 Sep 14;287(38):31633-40. doi: 10.1074/jbc.R112.349464. Epub 2012 Jul 20. J Biol Chem. 2012. PMID: 22822066 Free PMC article. Review.
-
Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.PLoS One. 2012;7(4):e34918. doi: 10.1371/journal.pone.0034918. Epub 2012 Apr 18. PLoS One. 2012. PMID: 22529956 Free PMC article.
-
Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38.J Biol Chem. 2009 Oct 2;284(40):27629-36. doi: 10.1074/jbc.M109.030965. Epub 2009 Jul 28. J Biol Chem. 2009. PMID: 19640843 Free PMC article.
-
A single residue at the active site of CD38 determines its NAD cyclizing and hydrolyzing activities.J Biol Chem. 2001 Apr 13;276(15):12169-73. doi: 10.1074/jbc.M011299200. Epub 2001 Jan 22. J Biol Chem. 2001. PMID: 11278881
-
ADP-ribosyl cyclase and CD38. Multi-functional enzymes in Ca+2 signaling.Adv Exp Med Biol. 1997;419:411-9. Adv Exp Med Biol. 1997. PMID: 9193683 Review.
Cited by
-
A Cell-Permeant Mimetic of NMN Activates SARM1 to Produce Cyclic ADP-Ribose and Induce Non-apoptotic Cell Death.iScience. 2019 May 31;15:452-466. doi: 10.1016/j.isci.2019.05.001. Epub 2019 May 4. iScience. 2019. PMID: 31128467 Free PMC article.
-
Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization.J Biol Chem. 2012 Sep 14;287(38):31633-40. doi: 10.1074/jbc.R112.349464. Epub 2012 Jul 20. J Biol Chem. 2012. PMID: 22822066 Free PMC article. Review.
-
Resolving the topological enigma in Ca2+ signaling by cyclic ADP-ribose and NAADP.J Biol Chem. 2019 Dec 27;294(52):19831-19843. doi: 10.1074/jbc.REV119.009635. Epub 2019 Oct 31. J Biol Chem. 2019. PMID: 31672920 Free PMC article. Review.
-
Immuno-targeting the multifunctional CD38 using nanobody.Sci Rep. 2016 Jun 2;6:27055. doi: 10.1038/srep27055. Sci Rep. 2016. PMID: 27251573 Free PMC article.
-
Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.Protein Sci. 2016 Mar;25(3):650-61. doi: 10.1002/pro.2859. Epub 2016 Jan 12. Protein Sci. 2016. PMID: 26660500 Free PMC article.
References
-
- Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, Aydin S. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–86. - PubMed
-
- Ferrero E, Malavasi F. The metamorphosis of a molecule: from soluble enzyme to the leukocyte receptor CD38. J Leukoc Biol. 1999;65:151–61. - PubMed
-
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. - PubMed
-
- Thornton PD, Fernandez C, Giustolisi GM, Morilla R, Atkinson S, A’Hern RP, Matutes E, Catovsky D. CD38 expression as a prognostic indicator in chronic lymphocytic leukaemia. Hematol J. 2004;5:145–51. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

