Fluorescence competition and optical melting measurements of RNA three-way multibranch loops provide a revised model for thermodynamic parameters
- PMID: 21133351
- PMCID: PMC3032278
- DOI: 10.1021/bi101470n
Fluorescence competition and optical melting measurements of RNA three-way multibranch loops provide a revised model for thermodynamic parameters
Abstract
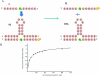
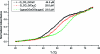
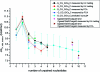
Three-way multibranch loops (junctions) are common in RNA secondary structures. Computer algorithms such as RNAstructure and MFOLD do not consider the identity of unpaired nucleotides in multibranch loops when predicting secondary structure. There is limited experimental data, however, to parametrize this aspect of these algorithms. In this study, UV optical melting and a fluorescence competition assay are used to measure stabilities of multibranch loops containing up to five unpaired adenosines or uridines or a loop E motif. These results provide a test of our understanding of the factors affecting multibranch loop stability and provide revised parameters for predicting stability. The results should help to improve predictions of RNA secondary structure.
Figures

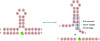
Similar articles
-
Experimentally derived nearest-neighbor parameters for the stability of RNA three- and four-way multibranch loops.Biochemistry. 2002 Jan 22;41(3):869-80. doi: 10.1021/bi011441d. Biochemistry. 2002. PMID: 11790109
-
Thermodynamics of three-way multibranch loops in RNA.Biochemistry. 2001 Jun 12;40(23):6971-81. doi: 10.1021/bi0029548. Biochemistry. 2001. PMID: 11389613
-
Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops.Biochemistry. 1991 Jul 2;30(26):6428-36. doi: 10.1021/bi00240a013. Biochemistry. 1991. PMID: 1711369
-
The energetics of small internal loops in RNA.Biopolymers. 1999-2000;52(4):157-67. doi: 10.1002/1097-0282(1999)52:4<157::AID-BIP1001>3.0.CO;2-E. Biopolymers. 1999. PMID: 11295748 Review.
-
Thermodynamic studies of RNA stability.J Biomol Struct Dyn. 1984 Mar;1(5):1229-42. doi: 10.1080/07391102.1984.10507514. J Biomol Struct Dyn. 1984. PMID: 6086053 Review.
Cited by
-
Bridging the gap between in vitro and in vivo RNA folding.Q Rev Biophys. 2016 Jan;49:e10. doi: 10.1017/S003358351600007X. Epub 2016 Jun 24. Q Rev Biophys. 2016. PMID: 27658939 Free PMC article.
-
The four ingredients of single-sequence RNA secondary structure prediction. A unifying perspective.RNA Biol. 2013 Jul;10(7):1185-96. doi: 10.4161/rna.24971. Epub 2013 May 10. RNA Biol. 2013. PMID: 23695796 Free PMC article.
-
Challenges with Simulating Modified RNA: Insights into Role and Reciprocity of Experimental and Computational Approaches.Genes (Basel). 2022 Mar 18;13(3):540. doi: 10.3390/genes13030540. Genes (Basel). 2022. PMID: 35328093 Free PMC article. Review.
-
Blind tests of RNA nearest-neighbor energy prediction.Proc Natl Acad Sci U S A. 2016 Jul 26;113(30):8430-5. doi: 10.1073/pnas.1523335113. Epub 2016 Jul 8. Proc Natl Acad Sci U S A. 2016. PMID: 27402765 Free PMC article.
-
Vfold2D-MC: A Physics-Based Hybrid Model for Predicting RNA Secondary Structure Folding.J Phys Chem B. 2021 Sep 16;125(36):10108-10118. doi: 10.1021/acs.jpcb.1c04731. Epub 2021 Sep 2. J Phys Chem B. 2021. PMID: 34473508 Free PMC article.
References
-
- Kim S. H.; Quigley G. J.; Suddath F. L.; McPherso A; Sneden D.; Kim J. J.; Weinzier J; Rich A. (1973) 3-dimensional structure of yeast phenylalanine transfer RNA: folding of polynucleotide chain. Science 179, 285–288. - PubMed
-
- Yusupov M. M.; Yusupova G. Z.; Baucom A.; Lieberman K.; Earnest T. N.; Cate J. H. D.; Noller H. F. (2001) Crystal structure of the ribosome at 5.5 angstrom resolution. Science 292, 883–896. - PubMed
-
- Leontis N. B.; Westhof E. (1998) A common motif organizes the structure of multi-helix loops in 16 and 23 S ribosomal RNAs. J. Mol. Biol. 283, 571–583. - PubMed
-
- Walter F.; Murchie A. I. H.; Lilley D. M. J. (1998) Folding of the four-way RNA junction of the hairpin ribozyme. Biochemistry 37, 17629–17636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

