DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo
- PMID: 21131571
- PMCID: PMC3009821
- DOI: 10.1073/pnas.1016382107
DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo
Abstract
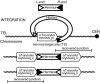
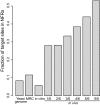
Transposons are mobile genetic elements that are an important source of genetic variation and are useful tools for genome engineering, mutagenesis screens, and vectors for transgenesis including gene therapy. We have used second-generation sequencing to analyze ≈2 × 10(5) unique de novo transposon insertion sites of the transposon Hermes in the Saccharomyces cerevisiae genome from both in vitro transposition reactions by using purified yeast genomic DNA, to better characterize intrinsic sequence specificity, and sites recovered from in vivo transposition events, to characterize the effect of intracellular factors such as chromatin on target site selection. We find that Hermes transposon targeting in vivo is profoundly affected by chromatin structure: The subset of genome-wide target sites used in vivo is strongly associated (P < 2e-16 by Fisher's exact test) with nucleosome-free chromatin. Our characterization of the insertion site preferences of Hermes not only assists in the future use of this transposon as a molecular biology tool but also establishes methods to more fully determine targeting mechanisms of other transposons. We have also discovered a long-range sequence motif that defines S. cerevisiae nucleosome-free regions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Genome-Wide Analysis of Nucleosome Positions, Occupancy, and Accessibility in Yeast: Nucleosome Mapping, High-Resolution Histone ChIP, and NCAM.Curr Protoc Mol Biol. 2014 Oct 1;108:21.28.1-21.28.16. doi: 10.1002/0471142727.mb2128s108. Curr Protoc Mol Biol. 2014. PMID: 25271716 Free PMC article. Review.
-
Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots.Genome Res. 2012 Apr;22(4):693-703. doi: 10.1101/gr.129460.111. Epub 2012 Jan 4. Genome Res. 2012. PMID: 22219510 Free PMC article.
-
A genomic code for nucleosome positioning.Nature. 2006 Aug 17;442(7104):772-8. doi: 10.1038/nature04979. Epub 2006 Jul 19. Nature. 2006. PMID: 16862119 Free PMC article.
-
Multipurpose transposon insertion libraries for large-scale analysis of gene function in yeast.Methods Mol Biol. 2008;416:117-29. doi: 10.1007/978-1-59745-321-9_8. Methods Mol Biol. 2008. PMID: 18392964
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
Cited by
-
Integration profiling of gene function with dense maps of transposon integration.Genetics. 2013 Oct;195(2):599-609. doi: 10.1534/genetics.113.152744. Epub 2013 Jul 26. Genetics. 2013. PMID: 23893486 Free PMC article.
-
A hyperactive transposase of the maize transposable element activator (Ac).Genetics. 2012 Jul;191(3):747-56. doi: 10.1534/genetics.112.139642. Epub 2012 May 4. Genetics. 2012. PMID: 22562933 Free PMC article.
-
"Calling cards" for DNA-binding proteins in mammalian cells.Genetics. 2012 Mar;190(3):941-9. doi: 10.1534/genetics.111.137315. Epub 2012 Jan 3. Genetics. 2012. PMID: 22214611 Free PMC article.
-
Epigenetic control of mobile DNA as an interface between experience and genome change.Front Genet. 2014 Apr 25;5:87. doi: 10.3389/fgene.2014.00087. eCollection 2014. Front Genet. 2014. PMID: 24795749 Free PMC article. Review.
-
Integration site selection by retroviruses and transposable elements in eukaryotes.Nat Rev Genet. 2017 May;18(5):292-308. doi: 10.1038/nrg.2017.7. Epub 2017 Mar 13. Nat Rev Genet. 2017. PMID: 28286338 Review.
References
-
- VandenDriessche T, Chuah MK. Moving gene therapy forward with mobile DNA. Hum Gene Ther. 2009;20:1559–1561. - PubMed
-
- Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. - PubMed
-
- Chalker DL, Sandmeyer SB. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

