A structural analysis of M protein in coronavirus assembly and morphology
- PMID: 21130884
- PMCID: PMC4486061
- DOI: 10.1016/j.jsb.2010.11.021
A structural analysis of M protein in coronavirus assembly and morphology
Abstract
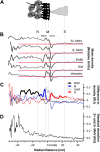
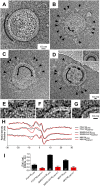
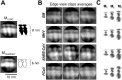
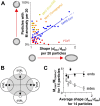
The M protein of coronavirus plays a central role in virus assembly, turning cellular membranes into workshops where virus and host factors come together to make new virus particles. We investigated how M structure and organization is related to virus shape and size using cryo-electron microscopy, tomography and statistical analysis. We present evidence that suggests M can adopt two conformations and that membrane curvature is regulated by one M conformer. Elongated M protein is associated with rigidity, clusters of spikes and a relatively narrow range of membrane curvature. In contrast, compact M protein is associated with flexibility and low spike density. Analysis of several types of virus-like particles and virions revealed that S protein, N protein and genomic RNA each help to regulate virion size and variation, presumably through interactions with M. These findings provide insight into how M protein functions to promote virus assembly.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Structural Analysis of the Roles of Influenza A Virus Membrane-Associated Proteins in Assembly and Morphology.J Virol. 2015 Sep;89(17):8957-66. doi: 10.1128/JVI.00592-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085153 Free PMC article.
-
Promotion of virus assembly and organization by the measles virus matrix protein.Nat Commun. 2018 Apr 30;9(1):1736. doi: 10.1038/s41467-018-04058-2. Nat Commun. 2018. PMID: 29712906 Free PMC article.
-
Analyses of Coronavirus Assembly Interactions with Interspecies Membrane and Nucleocapsid Protein Chimeras.J Virol. 2016 Apr 14;90(9):4357-4368. doi: 10.1128/JVI.03212-15. Print 2016 May. J Virol. 2016. PMID: 26889024 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Structure and organization of paramyxovirus particles.Curr Opin Virol. 2017 Jun;24:105-114. doi: 10.1016/j.coviro.2017.05.004. Epub 2017 Jun 8. Curr Opin Virol. 2017. PMID: 28601688 Free PMC article. Review.
Cited by
-
Can trace element supplementations (Cu, Se, and Zn) enhance human immunity against COVID-19 and its new variants?Beni Suef Univ J Basic Appl Sci. 2021;10(1):33. doi: 10.1186/s43088-021-00123-w. Epub 2021 May 17. Beni Suef Univ J Basic Appl Sci. 2021. PMID: 34026905 Free PMC article. Review.
-
SARS-CoV-2 and Human Immunodeficiency Virus: Pathogen Pincer Attack.HIV AIDS (Auckl). 2021 Mar 31;13:361-375. doi: 10.2147/HIV.S300055. eCollection 2021. HIV AIDS (Auckl). 2021. PMID: 33833585 Free PMC article. Review.
-
The 2020 Pandemic: Current SARS-CoV-2 Vaccine Development.Front Immunol. 2020 Aug 19;11:1880. doi: 10.3389/fimmu.2020.01880. eCollection 2020. Front Immunol. 2020. PMID: 32973779 Free PMC article. Review.
-
The SARS-CoV-2 Nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein.bioRxiv [Preprint]. 2020 Jul 31:2020.07.30.228023. doi: 10.1101/2020.07.30.228023. bioRxiv. 2020. Update in: Nat Commun. 2021 Jan 21;12(1):502. doi: 10.1038/s41467-020-20768-y PMID: 32766587 Free PMC article. Updated. Preprint.
-
The percentages of SARS-CoV-2 protein similarity and identity with SARS-CoV and BatCoV RaTG13 proteins can be used as indicators of virus origin.J Proteins Proteom. 2021;12(2):81-91. doi: 10.1007/s42485-021-00060-3. Epub 2021 Apr 9. J Proteins Proteom. 2021. PMID: 33850392 Free PMC article.
References
-
- Albertini A.A., Wernimont A.K., Muziol T., Ravelli R.B., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. - PubMed
-
- Arpin N., Talbot P.J. Molecular characterization of the 229E strain of human coronavirus. Adv. Exp. Med. Biol. 1990;276:73–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR17573/RR/NCRR NIH HHS/United States
- AI-059799/AI/NIAID NIH HHS/United States
- AI-29984/AI/NIAID NIH HHS/United States
- R01 AI059799/AI/NIAID NIH HHS/United States
- MC_UP_A550_1029/MRC_/Medical Research Council/United Kingdom
- AI-72493/AI/NIAID NIH HHS/United States
- R21 AI029984/AI/NIAID NIH HHS/United States
- R01 AI029984/AI/NIAID NIH HHS/United States
- MC_UU_12014/7/MRC_/Medical Research Council/United Kingdom
- HHSN-266200400058C/PHS HHS/United States
- AI-25913/AI/NIAID NIH HHS/United States
- P41 RR017573/RR/NCRR NIH HHS/United States
- R01 AI072493/AI/NIAID NIH HHS/United States
- R01 AI025913/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

