Mice defective in p53 nuclear localization signal 1 exhibit exencephaly
- PMID: 21127973
- PMCID: PMC4234151
- DOI: 10.1007/s11248-010-9468-4
Mice defective in p53 nuclear localization signal 1 exhibit exencephaly
Abstract
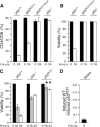
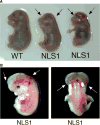
p53 is a major suppressor of human malignancy. The protein levels and activity are tightly regulated in cells. Early experiments identified nuclear localization signal 1 (NLS1) as a regulator of p53 localization. We have generated mice bearing a mutation in p53 ( NLS1 ), designated p53 ( NLS1 ). Our experiments confirm a role for NLS1 in regulating p53 function. Murine embryonic fibroblasts generated from homozygous p53 ( NLS1 ) animals are partially defective in cell cycle arrest and do not respond to inhibitory signals from oncogenic Ras. In addition, p53-dependent apoptosis is abrogated in thymocytes. Contrary to predicted results, fibroblasts from homozygous p53 ( NLS1 ) animals have a greater rate of proliferation than p53-null cells. In addition, p53 ( NLS1 ) cells are more resistant to UV-induced death. Surprisingly, the homozygous p53 ( NLS1 ) animals exhibit embryonic and peri-natal lethality, with a significant portion of the animals developing exencephaly. Thus, p53 ( NLS1/NLS1 ) embryos exhibit a reduced viability relative to p53-null mice. These studies indicate that the NLS1 is a major regulator of p53 activity in vivo.
Conflict of interest statement
Figures
Similar articles
-
Neural tube development requires the cooperation of p53- and Gadd45a-associated pathways.Birth Defects Res A Clin Mol Teratol. 2006 Feb;76(2):129-32. doi: 10.1002/bdra.20217. Birth Defects Res A Clin Mol Teratol. 2006. PMID: 16470852
-
Mutations in Lyar and p53 are synergistically lethal in female mice.Birth Defects Res A Clin Mol Teratol. 2012 Sep;94(9):729-37. doi: 10.1002/bdra.23048. Epub 2012 Jul 19. Birth Defects Res A Clin Mol Teratol. 2012. PMID: 22815056
-
An intragenic deletion of nuclear localization signal-1 of p53 tumor suppressor gene results in loss of apoptosis in murine fibroblasts.Cancer Lett. 1999 Dec 1;147(1-2):101-8. doi: 10.1016/s0304-3835(99)00283-9. Cancer Lett. 1999. PMID: 10660095
-
[Relationships between p53 induction, cell cycle arrest and survival of normal human fibroblasts following DNA damage].Bull Cancer. 1997 Nov;84(11):1007-16. Bull Cancer. 1997. PMID: 9536982 Review. French.
-
p53 in embryonic development: maintaining a fine balance.Cell Mol Life Sci. 1999 Jan;55(1):38-47. doi: 10.1007/s000180050268. Cell Mol Life Sci. 1999. PMID: 10065150 Free PMC article. Review.
Cited by
-
The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation.Cell Death Differ. 2017 Jun;24(6):1063-1078. doi: 10.1038/cdd.2017.54. Epub 2017 Apr 21. Cell Death Differ. 2017. PMID: 28430184 Free PMC article.
References
-
- Addison C, Jenkins JR, Sturzbecher HW. The p53 nuclear localisation signal is structurally linked to a p34cdc2 kinase motif. Oncogene. 1990;5:423–426. - PubMed
-
- Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res. 2007;67:11696–11703. - PubMed
-
- Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5:931–936. - PubMed
-
- Attardi LD, de Vries A, Jacks T. Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene. 2004;23:973–980. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

