Target genes of the largest human SWI/SNF complex subunit control cell growth
- PMID: 21118156
- PMCID: PMC4090146
- DOI: 10.1042/BJ20101358
Target genes of the largest human SWI/SNF complex subunit control cell growth
Abstract
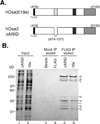
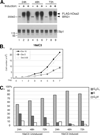
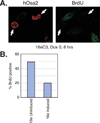
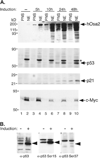
The largest subunit of the mammalian SWI/SNF-A or BAF (BRG1-associated factor) chromatin-remodelling complex is encoded by two related cDNAs hOsa1/BAF250a and hOsa2/BAF250b that are unique to the BAF complex and absent in the related PBAF (Polybromo BAF). hOsa/BAF250 has been shown to interact with transcriptional activators and bind to DNA suggesting that it acts to target the remodelling complex to chromatin. To better understand the functions of hOsa2, we established inducible stable HeLa cell lines over-expressing FLAG-hOsa2 or a derivative lacking the ARID (AT-rich interactive domain) DNA-binding domain. Immunopurification of complexes containing hOsa2 that was followed by mass spectrometry and immunoblotting demonstrated the presence of BRG1 and known BAFs, but not hOsa1 or hBRM. Deletion of the ARID did not compromise the integrity of the complex. Induction of hOsa2 expression caused impaired cell growth and accumulation of cells in the G0/G1 cell cycle phase. Elevated levels of the p53 and p21 proteins were detected in these cells while c-Myc mRNA and protein levels were found to decrease. Chromatin immunoprecipitation and reporter assays suggested that hOsa2 had a direct effect on c-myc and p21 promoter activity. Thus hOsa2 plays an important role in controlling genes regulating the cell cycle.
Figures

Similar articles
-
Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors.J Biol Chem. 2002 Nov 1;277(44):41674-85. doi: 10.1074/jbc.M205961200. Epub 2002 Aug 27. J Biol Chem. 2002. PMID: 12200431
-
Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes.J Biol Chem. 2018 Mar 16;293(11):3892-3903. doi: 10.1074/jbc.RA117.001065. Epub 2018 Jan 26. J Biol Chem. 2018. PMID: 29374058 Free PMC article.
-
Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes.J Cell Biochem. 2009 Oct 15;108(3):565-76. doi: 10.1002/jcb.22288. J Cell Biochem. 2009. PMID: 19650111
-
The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells.Oncogene. 2009 Jul 9;28(27):2492-501. doi: 10.1038/onc.2009.121. Epub 2009 May 18. Oncogene. 2009. PMID: 19448667 Free PMC article.
-
BAFfling pathologies: Alterations of BAF complexes in cancer.Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review.
Cited by
-
ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers.Cancer Res. 2011 Nov 1;71(21):6718-27. doi: 10.1158/0008-5472.CAN-11-1562. Epub 2011 Sep 7. Cancer Res. 2011. PMID: 21900401 Free PMC article.
-
Inactivation of chromatin remodeling factors sensitizes cells to selective cytotoxic stress.Biologics. 2014 Nov 14;8:269-80. doi: 10.2147/BTT.S67046. eCollection 2014. Biologics. 2014. PMID: 25484574 Free PMC article.
-
SWI/SNF chromatin remodeling complex: a new cofactor in reprogramming.Stem Cell Rev Rep. 2012 Mar;8(1):128-36. doi: 10.1007/s12015-011-9285-z. Stem Cell Rev Rep. 2012. PMID: 21655945 Review.
-
SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes.PLoS One. 2012;7(3):e33834. doi: 10.1371/journal.pone.0033834. Epub 2012 Mar 19. PLoS One. 2012. PMID: 22442726 Free PMC article.
-
CRISPR/Cas9-Mediated Gene Knockout of ARID1A Promotes Primary Progesterone Resistance by Downregulating Progesterone Receptor B in Endometrial Cancer Cells.Oncol Res. 2019 Sep 23;27(9):1051-1060. doi: 10.3727/096504019X15561873320465. Epub 2019 May 9. Oncol Res. 2019. PMID: 31072420 Free PMC article.
References
-
- Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J. Cell. Biochem. 2004;91:1087–1098. - PubMed
-
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. BioEssays. 2003;25:1192–1200. - PubMed
-
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. - PubMed
-
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. - PubMed
-
- Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

