TDP-43 toxicity in yeast
- PMID: 21115123
- PMCID: PMC3073690
- DOI: 10.1016/j.ymeth.2010.11.006
TDP-43 toxicity in yeast
Abstract
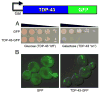
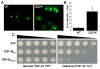
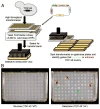
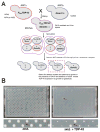
The budding yeast Saccharomyces cerevisiae is an emerging tool for investigating the molecular pathways that underpin several human neurodegenerative disorders associated with protein misfolding. Amyotrophic lateral sclerosis (ALS) is a devastating adult onset neurodegenerative disease primarily affecting motor neurons. The protein TDP-43 has recently been demonstrated to play an important role in the disease, however, the mechanisms by which TDP-43 contributes to pathogenesis are unclear. To explore the mechanistic details that result in aberrant accumulation of TDP-43 and to discover potential strategies for therapeutic intervention, we employed a yeast TDP-43 proteinopathy model system. These studies allowed us to determine the regions of TDP-43 required for aggregation and toxicity and to define the effects of ALS-linked mutant forms of TDP-43. We have also been able to harness the power of yeast genetics to identify potent modifiers of TDP-43 toxicity using high-throughput yeast genetic screens. Here, we describe the methods and approaches that we have used in order to gain insight into TDP-43 biology and its role in disease. These approaches are readily adaptable to other neurodegenerative disease proteins.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS.PLoS Biol. 2011 Apr;9(4):e1000614. doi: 10.1371/journal.pbio.1000614. Epub 2011 Apr 26. PLoS Biol. 2011. PMID: 21541367 Free PMC article.
-
TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration.Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3858-63. doi: 10.1073/pnas.0912417107. Epub 2010 Feb 3. Proc Natl Acad Sci U S A. 2010. PMID: 20133711 Free PMC article.
-
A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity.Proc Natl Acad Sci U S A. 2008 Apr 29;105(17):6439-44. doi: 10.1073/pnas.0802082105. Epub 2008 Apr 23. Proc Natl Acad Sci U S A. 2008. PMID: 18434538 Free PMC article.
-
[Clinical and pathological spectrum of TDP-43 associated ALS].Rinsho Shinkeigaku. 2010 Nov;50(11):940-2. doi: 10.5692/clinicalneurol.50.940. Rinsho Shinkeigaku. 2010. PMID: 21921519 Review. Japanese.
-
Possible concurrence of TDP-43, tau and other proteins in amyotrophic lateral sclerosis/frontotemporal lobar degeneration.Neuropathology. 2018 Feb;38(1):72-81. doi: 10.1111/neup.12428. Epub 2017 Sep 27. Neuropathology. 2018. PMID: 28960544 Review.
Cited by
-
RNA-binding proteins in neurodegeneration: mechanisms in aggregate.Genes Dev. 2017 Aug 1;31(15):1509-1528. doi: 10.1101/gad.304055.117. Genes Dev. 2017. PMID: 28912172 Free PMC article. Review.
-
ALS Yeast Models-Past Success Stories and New Opportunities.Front Mol Neurosci. 2018 Oct 30;11:394. doi: 10.3389/fnmol.2018.00394. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30425620 Free PMC article. Review.
-
Nuclear TAR DNA-binding protein 43: A new target for amyotrophic lateral sclerosis treatment.Neural Regen Res. 2013 Dec 15;8(35):3284-95. doi: 10.3969/j.issn.1673-5374.2013.35.003. Neural Regen Res. 2013. PMID: 25206650 Free PMC article.
-
Nucleolin Rescues TDP-43 Toxicity in Yeast and Human Cell Models.Front Cell Neurosci. 2021 Apr 12;15:625665. doi: 10.3389/fncel.2021.625665. eCollection 2021. Front Cell Neurosci. 2021. PMID: 33912014 Free PMC article.
-
Isolating potentiated Hsp104 variants using yeast proteinopathy models.J Vis Exp. 2014 Nov 11;(93):e52089. doi: 10.3791/52089. J Vis Exp. 2014. PMID: 25407485 Free PMC article.
References
-
- Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. Science. 1991;251:675–8. - PubMed
-
- Glenner GG, Wong CW. Biochem Biophys Res Commun. 1984;120:885–90. - PubMed
-
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Nature. 1997;388:839–40. - PubMed
-
- McKinley MP, Bolton DC, Prusiner SB. Cell. 1983;35:57–62. - PubMed
-
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Science. 1997;277:1990–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

