Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59
- PMID: 21106733
- PMCID: PMC3028919
- DOI: 10.1128/JVI.01709-10
Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59
Abstract
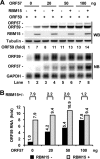
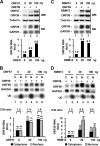
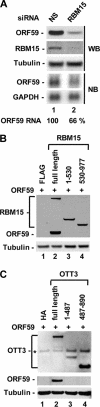
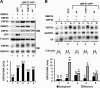
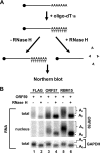
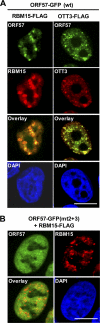
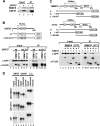
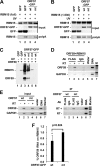
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes ORF57, which promotes the accumulation of specific KSHV mRNA targets, including ORF59 mRNA. We report that the cellular export NXF1 cofactors RBM15 and OTT3 participate in ORF57-enhanced expression of KSHV ORF59. We also found that ectopic expression of RBM15 or OTT3 augments ORF59 production in the absence of ORF57. While RBM15 promotes the accumulation of ORF59 RNA predominantly in the nucleus compared to the levels in the cytoplasm, we found that ORF57 shifted the nucleocytoplasmic balance by increasing ORF59 RNA accumulation in the cytoplasm more than in the nucleus. By promoting the accumulation of cytoplasmic ORF59 RNA, ORF57 offsets the nuclear RNA accumulation mediated by RBM15 by preventing nuclear ORF59 RNA from hyperpolyadenylation. ORF57 interacts directly with the RBM15 C-terminal portion containing the SPOC domain to reduce RBM15 binding to ORF59 RNA. Although ORF57 homologs Epstein-Barr virus (EBV) EB2, herpes simplex virus (HSV) ICP27, varicella-zoster virus (VZV) IE4/ORF4, and cytomegalovirus (CMV) UL69 also interact with RBM15 and OTT3, EBV EB2, which also promotes ORF59 expression, does not function like KSHV ORF57 to efficiently prevent RBM15-mediated nuclear accumulation of ORF59 RNA and RBM15's association with polyadenylated RNAs. Collectively, our data provide novel insight elucidating a molecular mechanism by which ORF57 promotes the expression of viral intronless genes.
Figures


Similar articles
-
Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57.Virology. 2010 Nov 25;407(2):206-12. doi: 10.1016/j.virol.2010.08.014. Epub 2010 Sep 9. Virology. 2010. PMID: 20828777 Free PMC article.
-
Multiple regions of Kaposi's sarcoma-associated herpesvirus ORF59 RNA are required for its expression mediated by viral ORF57 and cellular RBM15.Viruses. 2015 Feb 3;7(2):496-510. doi: 10.3390/v7020496. Viruses. 2015. PMID: 25690794 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export.J Virol. 2007 Sep;81(18):9990-8. doi: 10.1128/JVI.00896-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609285 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing.Front Biosci (Landmark Ed). 2009 Jan 1;14(4):1516-28. doi: 10.2741/3322. Front Biosci (Landmark Ed). 2009. PMID: 19273144 Free PMC article. Review.
-
The KSHV RNA regulator ORF57: target specificity and its role in the viral life cycle.Wiley Interdiscip Rev RNA. 2016 Mar-Apr;7(2):173-85. doi: 10.1002/wrna.1323. Epub 2016 Jan 14. Wiley Interdiscip Rev RNA. 2016. PMID: 26769399 Review.
Cited by
-
CLIP-seq to Identify KSHV ORF57-Binding RNA in Host B Cells.Curr Protoc Microbiol. 2016 May 6;41:1E.11.1-1E.11.18. doi: 10.1002/cpmc.3. Curr Protoc Microbiol. 2016. PMID: 27153386 Free PMC article.
-
Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl.Blood. 2015 Feb 5;125(6):941-8. doi: 10.1182/blood-2014-08-593392. Epub 2014 Dec 2. Blood. 2015. PMID: 25468569 Free PMC article.
-
Binding of cellular export factor REF/Aly by Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein is not required for efficient KSHV lytic replication.J Virol. 2012 Sep;86(18):9866-74. doi: 10.1128/JVI.01190-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761374 Free PMC article.
-
The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function.PLoS Pathog. 2018 Aug 10;14(8):e1007232. doi: 10.1371/journal.ppat.1007232. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30096191 Free PMC article.
-
SPOC domain proteins in health and disease.Genes Dev. 2023 Mar 1;37(5-6):140-170. doi: 10.1101/gad.350314.122. Epub 2023 Mar 16. Genes Dev. 2023. PMID: 36927757 Free PMC article. Review.
References
-
- Bello, L. J., et al. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

