Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins
- PMID: 21106652
- PMCID: PMC3004066
- DOI: 10.1261/rna.2498411
Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins
Abstract
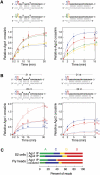
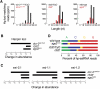
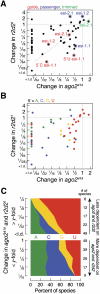
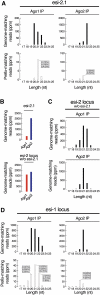
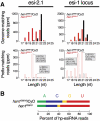
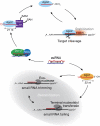
In flies, 22-23-nucleotide (nt) microRNA duplexes typically contain mismatches and begin with uridine, so they bind Argonaute1 (Ago1), whereas 21-nt siRNA duplexes are perfectly paired and begin with cytidine, promoting their loading into Ago2. A subset of Drosophila endogenous siRNAs-the hairpin-derived hp-esiRNAs-are born as mismatched duplexes that often begin with uridine. These would be predicted to load into Ago1, yet accumulate at steady-state bound to Ago2. In vitro, such hp-esiRNA duplexes assemble into Ago1. In vivo, they encounter complementary target mRNAs that trigger their tailing and trimming, causing Ago1-loaded hp-esiRNAs to be degraded. In contrast, Ago2-associated hp-esiRNAs are 2'-O-methyl modified at their 3' ends, protecting them from tailing and trimming. Consequently, the steady-state distribution of esiRNAs reflects not only their initial sorting between Ago1 and Ago2 according to their duplex structure, length, and first nucleotide, but also the targeted destruction of the single-stranded small RNAs after their loading into an Argonaute protein.
Figures

Similar articles
-
Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway.RNA. 2010 Jan;16(1):43-56. doi: 10.1261/rna.1972910. Epub 2009 Nov 16. RNA. 2010. PMID: 19917635 Free PMC article.
-
R2D2 organizes small regulatory RNA pathways in Drosophila.Mol Cell Biol. 2011 Feb;31(4):884-96. doi: 10.1128/MCB.01141-10. Epub 2010 Dec 6. Mol Cell Biol. 2011. PMID: 21135122 Free PMC article.
-
Sorting of Drosophila small silencing RNAs.Cell. 2007 Jul 27;130(2):299-308. doi: 10.1016/j.cell.2007.05.057. Cell. 2007. PMID: 17662944 Free PMC article.
-
Argonaute-mediated translational repression (and activation).Fly (Austin). 2009 Jul-Sep;3(3):204-6. Epub 2009 Jul 14. Fly (Austin). 2009. PMID: 19556851 Review.
-
Life of RISC: Formation, action, and degradation of RNA-induced silencing complex.Mol Cell. 2022 Jan 6;82(1):30-43. doi: 10.1016/j.molcel.2021.11.026. Epub 2021 Dec 22. Mol Cell. 2022. PMID: 34942118 Review.
Cited by
-
The Secret Garden of Neuronal circRNAs.Cells. 2020 Jul 31;9(8):1815. doi: 10.3390/cells9081815. Cells. 2020. PMID: 32751850 Free PMC article. Review.
-
Small RNA sorting: matchmaking for Argonautes.Nat Rev Genet. 2011 Jan;12(1):19-31. doi: 10.1038/nrg2916. Epub 2010 Nov 30. Nat Rev Genet. 2011. PMID: 21116305 Free PMC article. Review.
-
Diversifying microRNA sequence and function.Nat Rev Mol Cell Biol. 2013 Aug;14(8):475-88. doi: 10.1038/nrm3611. Epub 2013 Jun 26. Nat Rev Mol Cell Biol. 2013. PMID: 23800994 Review.
-
A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain.Cell. 2018 Jul 12;174(2):350-362.e17. doi: 10.1016/j.cell.2018.05.022. Epub 2018 Jun 7. Cell. 2018. PMID: 29887379 Free PMC article.
-
Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals.Genes Dev. 2013 Apr 1;27(7):778-92. doi: 10.1101/gad.211698.112. Epub 2013 Mar 27. Genes Dev. 2013. PMID: 23535236 Free PMC article.
References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 - PubMed
-
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236 doi: 10.1371/journal.pbio.0030236 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
