Novel role and mechanism of protein inhibitor of activated STAT1 in spatial learning
- PMID: 21102409
- PMCID: PMC3020114
- DOI: 10.1038/emboj.2010.290
Novel role and mechanism of protein inhibitor of activated STAT1 in spatial learning
Abstract
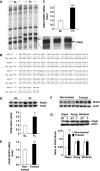
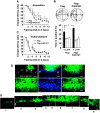
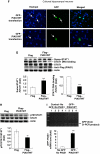
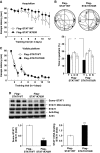
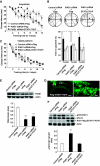
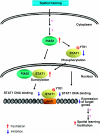
By using differential display PCR, we have previously identified 98 cDNA fragments from rat dorsal hippocampus, which are expressed differentially between the fast learners and slow learners from water-maze learning task. One cDNA fragment, which showed a higher expression level in fast learners, encodes the rat protein inhibitor of activated STAT1 (pias1) gene. Spatial training induced a significant increase in PIAS1 expression in rat hippocampus. Transient transfection of the wild-type (WT) PIAS1 plasmid to CA1 neurons facilitated, whereas transfection of PIAS1 siRNA impaired spatial learning in rats. Meanwhile, PIAS1WT increased STAT1 sumoylation, decreased STAT1 DNA binding and decreased STAT1 phosphorylation at Tyr-701 associated with spatial learning facilitation. But PIAS1 siRNA transfection produced an opposite effect on these measures associated with spatial learning impairment. Further, transfection of STAT1 sumoylation mutant impaired spatial acquisition, whereas transfection of STAT1 phosphorylation mutant blocked the impairing effect of PIAS1 siRNA on spatial learning. In this study, we first demonstrate the role of PIAS1 in spatial learning. Both posttranslational modifications (increased sumoylation and decreased phosphorylation) mediate the effect of PIAS1 on spatial learning facilitation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory.J Neurosci. 2014 Jul 16;34(29):9574-89. doi: 10.1523/JNEUROSCI.4302-13.2014. J Neurosci. 2014. PMID: 25031400 Free PMC article.
-
Phosphorylation of protein inhibitor of activated STAT1 (PIAS1) by MAPK-activated protein kinase-2 inhibits endothelial inflammation via increasing both PIAS1 transrepression and SUMO E3 ligase activity.Arterioscler Thromb Vasc Biol. 2013 Feb;33(2):321-9. doi: 10.1161/ATVBAHA.112.300619. Epub 2012 Nov 29. Arterioscler Thromb Vasc Biol. 2013. PMID: 23202365 Free PMC article.
-
NMDA receptor signaling mediates the expression of protein inhibitor of activated STAT1 (PIAS1) in rat hippocampus.Neuropharmacology. 2013 Feb;65:101-13. doi: 10.1016/j.neuropharm.2012.08.024. Epub 2012 Sep 7. Neuropharmacology. 2013. PMID: 22982248
-
Protein inhibitor of activated STAT1 Ser503 phosphorylation-mediated Elk-1 SUMOylation promotes neuronal survival in APP/PS1 mice.Br J Pharmacol. 2019 Jun;176(11):1793-1810. doi: 10.1111/bph.14656. Epub 2019 Apr 24. Br J Pharmacol. 2019. PMID: 30849179 Free PMC article.
-
Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes.Mol Cell. 2009 Sep 24;35(6):806-17. doi: 10.1016/j.molcel.2009.07.021. Mol Cell. 2009. PMID: 19782030
Cited by
-
Epigenetic regulation of HDAC1 SUMOylation as an endogenous neuroprotection against Aβ toxicity in a mouse model of Alzheimer's disease.Cell Death Differ. 2017 Apr;24(4):597-614. doi: 10.1038/cdd.2016.161. Epub 2017 Feb 10. Cell Death Differ. 2017. PMID: 28186506 Free PMC article.
-
Concerted type I interferon signaling in microglia and neural cells promotes memory impairment associated with amyloid β plaques.Immunity. 2022 May 10;55(5):879-894.e6. doi: 10.1016/j.immuni.2022.03.018. Epub 2022 Apr 19. Immunity. 2022. PMID: 35443157 Free PMC article.
-
STAT3 ameliorates cognitive deficits via regulation of NMDAR expression in an Alzheimer's disease animal model.Theranostics. 2021 Mar 13;11(11):5511-5524. doi: 10.7150/thno.56541. eCollection 2021. Theranostics. 2021. PMID: 33859760 Free PMC article.
-
Melatonin Induction of APP Intracellular Domain 50 SUMOylation Alleviates AD through Enhanced Transcriptional Activation and Aβ Degradation.Mol Ther. 2021 Jan 6;29(1):376-395. doi: 10.1016/j.ymthe.2020.09.003. Epub 2020 Sep 5. Mol Ther. 2021. PMID: 32950104 Free PMC article.
-
Smad4 SUMOylation is essential for memory formation through upregulation of the skeletal myopathy gene TPM2.BMC Biol. 2017 Nov 28;15(1):112. doi: 10.1186/s12915-017-0452-9. BMC Biol. 2017. PMID: 29183317 Free PMC article.
References
-
- Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA (1996) A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Human Gene Ther 7: 1947–1954 - PubMed
-
- Arvin B, Neville LF, Barone FC, Feuerstein GZ (1996) The role of inflammation and cytokines in brain injury. Neurosci Biobehav Rev 20: 445–452 - PubMed
-
- Burger C, Lopez MC, Feller JA, Baker HV, Muzyczka N, Mandel RJ (2007) Changes in transcription within the CA1 field of the hippocampus are associated with age-related spatial learning impairments. Neurobiol Learn Mem 87: 21–41 - PubMed
-
- Castellucci VF, Kennedy TE, Kandel ER, Goelet P (1988) A quantitative analysis of 2-D gels identifies proteins in which labeling is increased following long-term sensitization in Aplysia. Neuron 1: 321–328 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

