ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation
- PMID: 21095592
- PMCID: PMC3038635
- DOI: 10.1016/j.molcel.2010.11.002
ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation
Abstract
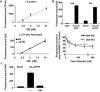
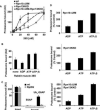
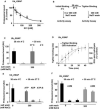
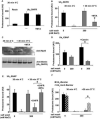
Eukaryotic cells target proteins for degradation by the 26S proteasome by attaching a ubiquitin chain. Using a rapid assay, we analyzed the initial binding of ubiquitinated proteins to purified 26S particles as an isolated process at 4°C. Subunits Rpn10 and Rpn13 contribute equally to the high-affinity binding of ubiquitin chains, but in their absence, ubiquitin conjugates bind to another site with 4-fold lower affinity. Conjugate binding is stimulated 2- to 4-fold by binding of ATP or the nonhydrolyzable analog, ATPγS (but not ADP), to the 19S ATPases. Following this initial, reversible association, ubiquitin conjugates at 37°C become more tightly bound through a step that requires ATP hydrolysis and a loosely folded domain on the protein, but appears independent of ubiquitin. Unfolded or loosely folded polypeptides can inhibit this tighter binding. This commitment step precedes substrate deubiquitination and allows for selection of ubiquitinated proteins capable of being unfolded and efficiently degraded.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Structure and Function of the 26S Proteasome.Annu Rev Biochem. 2018 Jun 20;87:697-724. doi: 10.1146/annurev-biochem-062917-011931. Epub 2018 Apr 13. Annu Rev Biochem. 2018. PMID: 29652515 Free PMC article. Review.
-
The recognition of ubiquitinated proteins by the proteasome.Cell Mol Life Sci. 2016 Sep;73(18):3497-506. doi: 10.1007/s00018-016-2255-5. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137187 Free PMC article. Review.
-
ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle.Cell. 2005 May 20;121(4):553-565. doi: 10.1016/j.cell.2005.03.028. Cell. 2005. Retraction in: Cell. 2018 Apr 19;173(3):804. doi: 10.1016/j.cell.2018.04.009. PMID: 15907469 Retracted.
-
The 26S Proteasome Switches between ATP-Dependent and -Independent Mechanisms in Response to Substrate Ubiquitination.Biomolecules. 2022 May 26;12(6):750. doi: 10.3390/biom12060750. Biomolecules. 2022. PMID: 35740875 Free PMC article.
-
Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening.Mol Cell. 2009 Dec 11;36(5):794-804. doi: 10.1016/j.molcel.2009.11.015. Mol Cell. 2009. PMID: 20005843 Free PMC article.
Cited by
-
Structure and Function of the 26S Proteasome.Annu Rev Biochem. 2018 Jun 20;87:697-724. doi: 10.1146/annurev-biochem-062917-011931. Epub 2018 Apr 13. Annu Rev Biochem. 2018. PMID: 29652515 Free PMC article. Review.
-
Regulating protein breakdown through proteasome phosphorylation.Biochem J. 2017 Sep 24;474(19):3355-3371. doi: 10.1042/BCJ20160809. Biochem J. 2017. PMID: 28947610 Free PMC article. Review.
-
AAA-ATPases in Protein Degradation.Front Mol Biosci. 2017 Jun 20;4:42. doi: 10.3389/fmolb.2017.00042. eCollection 2017. Front Mol Biosci. 2017. PMID: 28676851 Free PMC article. Review.
-
The Structural Role of RPN10 in the 26S Proteasome and an RPN2-Binding Residue on RPN13 Are Functionally Important in Arabidopsis.Int J Mol Sci. 2024 Oct 30;25(21):11650. doi: 10.3390/ijms252111650. Int J Mol Sci. 2024. PMID: 39519207 Free PMC article.
-
The recognition of ubiquitinated proteins by the proteasome.Cell Mol Life Sci. 2016 Sep;73(18):3497-506. doi: 10.1007/s00018-016-2255-5. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137187 Free PMC article. Review.
References
-
- Bech-Otschir D, Helfrich A, Enenkel C, Consiglieri G, Seeger M, Holzhutter HG, Dahlmann B, Kloetzel PM. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol. 2009;16:219–225. - PubMed
-
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. - PubMed
-
- Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol. 2009;394:732–746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

