Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling
- PMID: 21085662
- PMCID: PMC2978095
- DOI: 10.1371/journal.pone.0013927
Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling
Abstract
Type I interferons (IFNs) function as the first line of defense against viral infections by modulating cell growth, establishing an antiviral state and influencing the activation of various immune cells. Viruses such as influenza have developed mechanisms to evade this defense mechanism and during infection with influenza A viruses, the non-structural protein 1 (NS1) encoded by the virus genome suppresses induction of IFNs-α/β. Here we show that expression of avian H5N1 NS1 in HeLa cells leads to a block in IFN signaling. H5N1 NS1 reduces IFN-inducible tyrosine phosphorylation of STAT1, STAT2 and STAT3 and inhibits the nuclear translocation of phospho-STAT2 and the formation of IFN-inducible STAT1:1-, STAT1:3- and STAT3:3- DNA complexes. Inhibition of IFN-inducible STAT signaling by NS1 in HeLa cells is, in part, a consequence of NS1-mediated inhibition of expression of the IFN receptor subunit, IFNAR1. In support of this NS1-mediated inhibition, we observed a reduction in expression of ifnar1 in ex vivo human non-tumor lung tissues infected with H5N1 and H1N1 viruses. Moreover, H1N1 and H5N1 virus infection of human monocyte-derived macrophages led to inhibition of both ifnar1 and ifnar2 expression. In addition, NS1 expression induces up-regulation of the JAK/STAT inhibitors, SOCS1 and SOCS3. By contrast, treatment of ex vivo human lung tissues with IFN-α results in the up-regulation of a number of IFN-stimulated genes and inhibits both H5N1 and H1N1 virus replication. The data suggest that NS1 can directly interfere with IFN signaling to enhance viral replication, but that treatment with IFN can nevertheless override these inhibitory effects to block H5N1 and H1N1 virus infections.
Conflict of interest statement
Figures
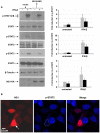
 ) or HA-tagged NS1 plasmid (▪) were left untreated (−) or treated (+) with IFN-β (1×103 U/mL) for 15 mins, 24 hr post-transfection. Cells were harvested, lysates were resolved by SDS-PAGE and immunoblotted with the indicated anti-phospho-STAT1, anti-phospho-STAT2, anti-phospho-STAT3 and anti-HA(NS1) antibodies. Membranes were stripped and reprobed with anti-STAT1, anti-STAT2, anti-STAT3 and anti-β-tubulin antibodies as loading controls. Relative fold induction of phosphorylated STAT proteins was calculated using signal intensity of phospho-STATs over total STATs and normalized with untreated, vector transfected cells. The data plots are representative of three independent experiments. B) HeLa cells transfected with HA-tagged NS1 plasmid were treated with IFN-β (1×103 U/mL) for 15 mins. Cells were then fixed and stained for HA (red) and phospho-STAT2 (blue), and analyzed by confocal microscopy as described in Materials and Methods. The white arrow identifies the nuclear predominance of NS1and the broken line in the middle panel defines the nucleus showing reduced phospho-STAT2. Data are representative of two independent experiments.
) or HA-tagged NS1 plasmid (▪) were left untreated (−) or treated (+) with IFN-β (1×103 U/mL) for 15 mins, 24 hr post-transfection. Cells were harvested, lysates were resolved by SDS-PAGE and immunoblotted with the indicated anti-phospho-STAT1, anti-phospho-STAT2, anti-phospho-STAT3 and anti-HA(NS1) antibodies. Membranes were stripped and reprobed with anti-STAT1, anti-STAT2, anti-STAT3 and anti-β-tubulin antibodies as loading controls. Relative fold induction of phosphorylated STAT proteins was calculated using signal intensity of phospho-STATs over total STATs and normalized with untreated, vector transfected cells. The data plots are representative of three independent experiments. B) HeLa cells transfected with HA-tagged NS1 plasmid were treated with IFN-β (1×103 U/mL) for 15 mins. Cells were then fixed and stained for HA (red) and phospho-STAT2 (blue), and analyzed by confocal microscopy as described in Materials and Methods. The white arrow identifies the nuclear predominance of NS1and the broken line in the middle panel defines the nucleus showing reduced phospho-STAT2. Data are representative of two independent experiments.
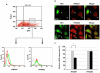
 ) or GFP vector containing HA-tagged NS1 (▪). 24 hr post-transfection, GFP positive cells were FACS sorted, RNA extracted and cDNA synthesized. Ifnar1, ifnar2 and β-actin gene expression were analyzed by RT-PCR. Gene expression was calculated relative to β-actin gene expression and normalized to cells transfected with GFP vector alone. Data are representative of two independent experiments. Significant differences (asterisk) were determined by Student's t-test (p<0.05).
) or GFP vector containing HA-tagged NS1 (▪). 24 hr post-transfection, GFP positive cells were FACS sorted, RNA extracted and cDNA synthesized. Ifnar1, ifnar2 and β-actin gene expression were analyzed by RT-PCR. Gene expression was calculated relative to β-actin gene expression and normalized to cells transfected with GFP vector alone. Data are representative of two independent experiments. Significant differences (asterisk) were determined by Student's t-test (p<0.05).
 ) expression was assayed by real-time PCR. Data shown are fold induction of gene expression relative to mock-infected control after normalizing to β-actin in each sample. Representative data of duplicate experiments with means of triplicate assays are shown; D) Human lung explant tissue was either mock-infected (PBS) or infected with A/HK/483/97 H5N1 or A/HK/54/98 H1N1 influenza A viruses, as described in Materials & Methods. 18 hr post-infection tissue was processed to extract RNA. cDNA was synthesized and expression of ifnar1, ifnar2 and β-actin gene expression was measured by RT-PCR analysis. Gene expression was calculated relative to β-actin gene expression and normalized to mock infected tissues. Data are representative of two independent experiments. Significant differences (asterisk) were determined by Student's t-test (p<0.05).
) expression was assayed by real-time PCR. Data shown are fold induction of gene expression relative to mock-infected control after normalizing to β-actin in each sample. Representative data of duplicate experiments with means of triplicate assays are shown; D) Human lung explant tissue was either mock-infected (PBS) or infected with A/HK/483/97 H5N1 or A/HK/54/98 H1N1 influenza A viruses, as described in Materials & Methods. 18 hr post-infection tissue was processed to extract RNA. cDNA was synthesized and expression of ifnar1, ifnar2 and β-actin gene expression was measured by RT-PCR analysis. Gene expression was calculated relative to β-actin gene expression and normalized to mock infected tissues. Data are representative of two independent experiments. Significant differences (asterisk) were determined by Student's t-test (p<0.05).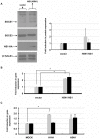
 , SOCS3 ▪). Data are representative of two independent experiments. B) HeLa cells were transfected with either GFP vector alone (
, SOCS3 ▪). Data are representative of two independent experiments. B) HeLa cells were transfected with either GFP vector alone ( ) or GFP vector containing HA-tagged NS1 (▪). 24 hr post-transfection, GFP+ cells were sorted, RNA extracted and cDNA synthesized. Gene expression of socs1 (
) or GFP vector containing HA-tagged NS1 (▪). 24 hr post-transfection, GFP+ cells were sorted, RNA extracted and cDNA synthesized. Gene expression of socs1 ( ), socs3 (▪) and β-actin gene expression were analyzed by RT-PCR. Gene expression was calculated relative to β-actin gene expression and normalized to cells transfected with GFP vector alone. Data are representative of two independent experiments. C) RNA from human lung tissue either mock-infected (PBS) or infected with A/HK/54/98 H1N1 or H5N1 influenza A viruses was collected 18 hr post-infection. cDNA was synthesized and expression of socs1(▪), socs3(▪) and β-actin gene expression was measured by RT-PCR analysis. Gene expression was calculated relative to β-actin gene expression and normalized to mock infected cells. Data are representative of two independent experiments. Significant differences (asterisk) were determined by Student's t-test (p<0.05).
), socs3 (▪) and β-actin gene expression were analyzed by RT-PCR. Gene expression was calculated relative to β-actin gene expression and normalized to cells transfected with GFP vector alone. Data are representative of two independent experiments. C) RNA from human lung tissue either mock-infected (PBS) or infected with A/HK/54/98 H1N1 or H5N1 influenza A viruses was collected 18 hr post-infection. cDNA was synthesized and expression of socs1(▪), socs3(▪) and β-actin gene expression was measured by RT-PCR analysis. Gene expression was calculated relative to β-actin gene expression and normalized to mock infected cells. Data are representative of two independent experiments. Significant differences (asterisk) were determined by Student's t-test (p<0.05).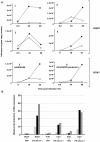
 ), isg15 (▪), 2′5′-oas (
), isg15 (▪), 2′5′-oas ( ), and β-actin, was measured by RT-PCR analysis at 18 hr post-infection with H5N1 or A/HK/54/98 H1N1 for explant 5. Data are representative of two independent experiments and normalized to mock infected controls.
), and β-actin, was measured by RT-PCR analysis at 18 hr post-infection with H5N1 or A/HK/54/98 H1N1 for explant 5. Data are representative of two independent experiments and normalized to mock infected controls.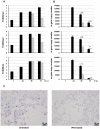
 ) for a further 48 hr. At the indicated times A) Viral titers (TCID50) and B) Influenza A m gene expression were measured; Significant differences were determined by Student t-test: * ∼ p<0.05; ** ∼ p<0.01. C) Thin sections of infected human lung explants, either untreated (i) or IFN-treated (ii) were stained for influenza A nucleoprotein (pink).
) for a further 48 hr. At the indicated times A) Viral titers (TCID50) and B) Influenza A m gene expression were measured; Significant differences were determined by Student t-test: * ∼ p<0.05; ** ∼ p<0.01. C) Thin sections of infected human lung explants, either untreated (i) or IFN-treated (ii) were stained for influenza A nucleoprotein (pink).Similar articles
-
A Conserved Residue, Tyrosine (Y) 84, in H5N1 Influenza A Virus NS1 Regulates IFN Signaling Responses to Enhance Viral Infection.Viruses. 2017 May 12;9(5):107. doi: 10.3390/v9050107. Viruses. 2017. PMID: 28498306 Free PMC article.
-
Influenza virus non‑structural protein 1 inhibits the production of interferon β of alveolar epithelial cells upon the infection of influenza A H1N1.Mol Med Rep. 2017 Oct;16(4):4553-4560. doi: 10.3892/mmr.2017.7138. Epub 2017 Aug 2. Mol Med Rep. 2017. PMID: 28765967 Free PMC article.
-
Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies.J Virol. 2015 Apr;89(8):4227-36. doi: 10.1128/JVI.00154-15. Epub 2015 Jan 28. J Virol. 2015. PMID: 25631085 Free PMC article.
-
Functions of the influenza A virus NS1 protein in antiviral defense.Curr Opin Virol. 2015 Jun;12:1-6. doi: 10.1016/j.coviro.2015.01.007. Epub 2015 Jan 29. Curr Opin Virol. 2015. PMID: 25638592 Free PMC article. Review.
-
The influenza virus NS1 protein as a therapeutic target.Antiviral Res. 2013 Sep;99(3):409-16. doi: 10.1016/j.antiviral.2013.06.005. Epub 2013 Jun 21. Antiviral Res. 2013. PMID: 23796981 Free PMC article. Review.
Cited by
-
An overview of the highly pathogenic H5N1 influenza virus.Virol Sin. 2013 Feb;28(1):3-15. doi: 10.1007/s12250-013-3294-9. Epub 2013 Jan 16. Virol Sin. 2013. PMID: 23325419 Free PMC article. Review.
-
Differential interferon responses to influenza A and B viruses in primary ferret respiratory epithelial cells.J Virol. 2024 Feb 20;98(2):e0149423. doi: 10.1128/jvi.01494-23. Epub 2024 Jan 31. J Virol. 2024. PMID: 38294251 Free PMC article.
-
Global virus outbreaks: Interferons as 1st responders.Semin Immunol. 2019 Jun;43:101300. doi: 10.1016/j.smim.2019.101300. Semin Immunol. 2019. PMID: 31771760 Free PMC article. Review.
-
Utilization of Aloe Compounds in Combatting Viral Diseases.Pharmaceuticals (Basel). 2022 May 13;15(5):599. doi: 10.3390/ph15050599. Pharmaceuticals (Basel). 2022. PMID: 35631425 Free PMC article. Review.
-
Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials.PLoS One. 2013 Aug 6;8(8):e70869. doi: 10.1371/journal.pone.0070869. Print 2013. PLoS One. 2013. PMID: 23940654 Free PMC article.
References
-
- Saito T, Gale M., Jr Principles of intracellular viral recognition. Curr Opin Immunol. 2007;19:17–23. - PubMed
-
- Hengel H, Koszinowski UH, Conzelmann KK. Viruses know it all: new insights into IFN networks. Trends Immunol. 2005;26:396–401. - PubMed
-
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. - PubMed
-
- Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, et al. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

