Structure and catalysis of acylaminoacyl peptidase: closed and open subunits of a dimer oligopeptidase
- PMID: 21084296
- PMCID: PMC3023495
- DOI: 10.1074/jbc.M110.169862
Structure and catalysis of acylaminoacyl peptidase: closed and open subunits of a dimer oligopeptidase
Abstract
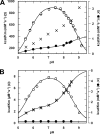
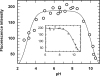
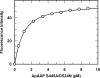
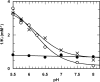
Acylaminoacyl peptidase from Aeropyrum pernix is a homodimer that belongs to the prolyl oligopeptidase family. The monomer subunit is composed of one hydrolase and one propeller domain. Previous crystal structure determinations revealed that the propeller domain obstructed the access of substrate to the active site of both subunits. Here we investigated the structure and the kinetics of two mutant enzymes in which the aspartic acid of the catalytic triad was changed to alanine or asparagine. Using different substrates, we have determined the pH dependence of specificity rate constants, the rate-limiting step of catalysis, and the binding of substrates and inhibitors. The catalysis considerably depended both on the kind of mutation and on the nature of the substrate. The results were interpreted in terms of alterations in the position of the catalytic histidine side chain as demonstrated with crystal structure determination of the native and two mutant structures (D524N and D524A). Unexpectedly, in the homodimeric structures, only one subunit displayed the closed form of the enzyme. The other subunit exhibited an open gate to the catalytic site, thus revealing the structural basis that controls the oligopeptidase activity. The open form of the native enzyme displayed the catalytic triad in a distorted, inactive state. The mutations affected the closed, active form of the enzyme, disrupting its catalytic triad. We concluded that the two forms are at equilibrium and the substrates bind by the conformational selection mechanism.
Figures

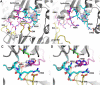
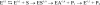
Similar articles
-
Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.Acta Crystallogr D Biol Crystallogr. 2015 Mar;71(Pt 3):461-72. doi: 10.1107/S1399004714026819. Epub 2015 Feb 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25760596
-
The acylaminoacyl peptidase from Aeropyrum pernix K1 thought to be an exopeptidase displays endopeptidase activity.J Mol Biol. 2007 Apr 27;368(2):509-20. doi: 10.1016/j.jmb.2007.02.025. Epub 2007 Feb 20. J Mol Biol. 2007. PMID: 17350041
-
Mechanisms of intramolecular communication in a hyperthermophilic acylaminoacyl peptidase: a molecular dynamics investigation.PLoS One. 2012;7(4):e35686. doi: 10.1371/journal.pone.0035686. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558199 Free PMC article.
-
The prolyl oligopeptidase family.Cell Mol Life Sci. 2002 Feb;59(2):349-62. doi: 10.1007/s00018-002-8427-5. Cell Mol Life Sci. 2002. PMID: 11915948 Free PMC article. Review.
-
Structure-function properties of prolyl oligopeptidase family enzymes.Cell Biochem Biophys. 2006;44(3):349-65. doi: 10.1385/CBB:44:3:349. Cell Biochem Biophys. 2006. PMID: 16679522 Review.
Cited by
-
Identification and characterisation of a novel acylpeptide hydrolase from Sulfolobus solfataricus: structural and functional insights.PLoS One. 2012;7(5):e37921. doi: 10.1371/journal.pone.0037921. Epub 2012 May 24. PLoS One. 2012. PMID: 22655081 Free PMC article.
-
Cryo-EM structure of acylpeptide hydrolase reveals substrate selection by multimerization and a multi-state serine-protease triad.Chem Sci. 2022 May 18;13(24):7132-7142. doi: 10.1039/d2sc02276a. eCollection 2022 Jun 22. Chem Sci. 2022. PMID: 35799812 Free PMC article.
-
Identification of oxidized protein hydrolase as a potential prodrug target in prostate cancer.BMC Cancer. 2014 Feb 10;14:77. doi: 10.1186/1471-2407-14-77. BMC Cancer. 2014. PMID: 24512522 Free PMC article.
-
Crystal structures of Trypanosoma brucei oligopeptidase B broaden the paradigm of catalytic regulation in prolyl oligopeptidase family enzymes.PLoS One. 2013 Nov 12;8(11):e79349. doi: 10.1371/journal.pone.0079349. eCollection 2013. PLoS One. 2013. PMID: 24265767 Free PMC article.
-
Novel Insights Into Leishmania (Viannia) braziliensis In Vitro Fitness Guided by Temperature Changes Along With Its Subtilisins and Oligopeptidase B.Front Cell Infect Microbiol. 2022 Apr 21;12:805106. doi: 10.3389/fcimb.2022.805106. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35531337 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

