The prion hypothesis: from biological anomaly to basic regulatory mechanism
- PMID: 21081963
- PMCID: PMC3003427
- DOI: 10.1038/nrm3007
The prion hypothesis: from biological anomaly to basic regulatory mechanism
Abstract
Prions are unusual proteinaceous infectious agents that are typically associated with a class of fatal degenerative diseases of the mammalian brain. However, the discovery of fungal prions, which are not associated with disease, suggests that we must now consider the effect of these factors on basic cellular physiology in a different light. Fungal prions are epigenetic determinants that can alter a range of cellular processes, including metabolism and gene expression pathways, and these changes can lead to a range of prion-associated phenotypes. The mechanistic similarities between prion propagation in mammals and fungi suggest that prions are not a biological anomaly but instead could be a newly appreciated and perhaps ubiquitous regulatory mechanism.
Figures

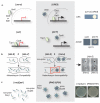
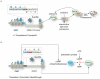
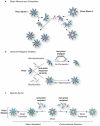
Comment in
-
Conformational conversion and prion disease.Nat Rev Mol Cell Biol. 2011 Apr;12(4):273; author reply 273. doi: 10.1038/nrm3007-c1. Nat Rev Mol Cell Biol. 2011. PMID: 21427768 No abstract available.
Similar articles
-
Fungal prions.Prog Mol Biol Transl Sci. 2012;107:417-56. doi: 10.1016/B978-0-12-385883-2.00007-2. Prog Mol Biol Transl Sci. 2012. PMID: 22482457 Review.
-
Amyloid formation of a yeast prion determinant.J Mol Neurosci. 2004;23(1-2):13-22. doi: 10.1385/JMN:23:1-2:013. J Mol Neurosci. 2004. PMID: 15126688 Review.
-
Is loss of function of the prion protein the cause of prion disorders?Trends Mol Med. 2003 Jun;9(6):237-43. doi: 10.1016/s1471-4914(03)00069-8. Trends Mol Med. 2003. PMID: 12829011
-
Propagating prions in fungi and mammals.Mol Cell. 2004 Jun 4;14(5):541-52. doi: 10.1016/j.molcel.2004.05.012. Mol Cell. 2004. PMID: 15175150 Review.
-
Molecular basis of cerebral neurodegeneration in prion diseases.FEBS J. 2007 Feb;274(3):606-11. doi: 10.1111/j.1742-4658.2007.05633.x. FEBS J. 2007. PMID: 17288549 Review.
Cited by
-
Functional diversification of hsp40: distinct j-protein functional requirements for two prions allow for chaperone-dependent prion selection.PLoS Genet. 2014 Jul 24;10(7):e1004510. doi: 10.1371/journal.pgen.1004510. eCollection 2014 Jul. PLoS Genet. 2014. PMID: 25058638 Free PMC article.
-
How and why to build a mathematical model: A case study using prion aggregation.J Biol Chem. 2020 Apr 10;295(15):5022-5035. doi: 10.1074/jbc.REV119.009851. Epub 2020 Jan 31. J Biol Chem. 2020. PMID: 32005659 Free PMC article. Review.
-
Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding.Nat Commun. 2014 Jul 15;5:4383. doi: 10.1038/ncomms5383. Nat Commun. 2014. PMID: 25023910 Free PMC article.
-
The expanding amyloid family: Structure, stability, function, and pathogenesis.Cell. 2021 Sep 16;184(19):4857-4873. doi: 10.1016/j.cell.2021.08.013. Cell. 2021. PMID: 34534463 Free PMC article. Review.
-
Yeast Short-Lived Actin-Associated Protein Forms a Metastable Prion in Response to Thermal Stress.Cell Rep. 2017 Jan 17;18(3):751-761. doi: 10.1016/j.celrep.2016.12.082. Cell Rep. 2017. PMID: 28099852 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

