A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3'-end formation
- PMID: 21078872
- PMCID: PMC3019983
- DOI: 10.1128/MCB.00943-10
A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3'-end formation
Abstract
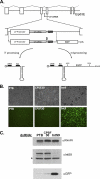
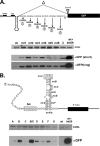
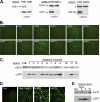
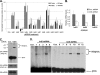
Proper gene expression relies on a class of ubiquitously expressed, uridine-rich small nuclear RNAs (snRNAs) transcribed by RNA polymerase II (RNAPII). Vertebrate snRNAs are transcribed from a unique promoter, which is required for proper 3'-end formation, and cleavage of the nascent transcript involves the activity of a poorly understood set of proteins called the Integrator complex. To examine 3'-end formation in Drosophila melanogaster, we developed a cell-based reporter that monitors aberrant 3'-end formation of snRNA through the gain in expression of green fluorescent protein (GFP). We used this reporter in Drosophila S2 cells to determine requirements for U7 snRNA 3'-end formation and found that processing was strongly dependent upon nucleotides located within the 3' stem-loop as well as sequences likely to comprise the Drosophila equivalent of the vertebrate 3' box. Substitution of the actin promoter for the snRNA promoter abolished proper 3'-end formation, demonstrating the conserved requirement for an snRNA promoter in Drosophila. We tested the requirement for all Drosophila Integrator subunits and found that Integrators 1, 4, 9, and 11 were essential for 3'-end formation and that Integrators 3 and 10 may be dispensable for processing. Depletion of cleavage and polyadenylation factors or of histone pre-mRNA processing factors did not affect U7 snRNA processing efficiency, demonstrating that the Integrator complex does not share components with the mRNA 3'-end processing machinery. Finally, flies harboring mutations in either Integrator 4 or 7 fail to complete development and accumulate significant levels of misprocessed snRNA in the larval stages.
Figures


Similar articles
-
An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3'-end formation.RNA. 2012 Dec;18(12):2148-56. doi: 10.1261/rna.035725.112. Epub 2012 Oct 24. RNA. 2012. PMID: 23097424 Free PMC article.
-
Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc).Crit Rev Biochem Mol Biol. 2011 Feb;46(1):11-26. doi: 10.3109/10409238.2010.518136. Epub 2010 Oct 6. Crit Rev Biochem Mol Biol. 2011. PMID: 20925482 Review.
-
Cloning and characterization of the Drosophila U7 small nuclear RNA.Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9422-7. doi: 10.1073/pnas.1533509100. Epub 2003 Jul 18. Proc Natl Acad Sci U S A. 2003. PMID: 12872004 Free PMC article.
-
U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis.RNA. 2006 Mar;12(3):396-409. doi: 10.1261/rna.2270406. RNA. 2006. PMID: 16495235 Free PMC article.
-
snRNA 3' end formation: the dawn of the Integrator complex.Biochem Soc Trans. 2010 Aug;38(4):1082-7. doi: 10.1042/BST0381082. Biochem Soc Trans. 2010. PMID: 20659008 Free PMC article. Review.
Cited by
-
Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex.J Biol Chem. 2013 Feb 15;288(7):4867-77. doi: 10.1074/jbc.M112.425892. Epub 2013 Jan 3. J Biol Chem. 2013. PMID: 23288851 Free PMC article.
-
The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3' ends.Genes Dev. 2015 Jul 15;29(14):1552-64. doi: 10.1101/gad.266973.115. Genes Dev. 2015. PMID: 26220997 Free PMC article.
-
Inositol hexakisphosphate is required for Integrator function.Nat Commun. 2022 Sep 30;13(1):5742. doi: 10.1038/s41467-022-33506-3. Nat Commun. 2022. PMID: 36180473 Free PMC article.
-
An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3'-end formation.RNA. 2012 Dec;18(12):2148-56. doi: 10.1261/rna.035725.112. Epub 2012 Oct 24. RNA. 2012. PMID: 23097424 Free PMC article.
-
The Integrator complex controls the termination of transcription at diverse classes of gene targets.Cell Res. 2015 Mar;25(3):288-305. doi: 10.1038/cr.2015.19. Epub 2015 Feb 13. Cell Res. 2015. PMID: 25675981 Free PMC article.
References
-
- Armknecht, S., et al. 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 392:55-73. - PubMed
-
- Baillat, D., et al. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123:265-276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
