RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1
- PMID: 21068368
- PMCID: PMC2996456
- DOI: 10.1073/pnas.1009231107
RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1
Abstract
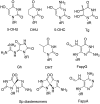
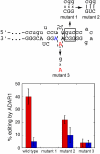
Editing of the pre-mRNA for the DNA repair enzyme NEIL1 causes a lysine to arginine change in the lesion recognition loop of the protein. The two forms of NEIL1 are shown here to have distinct enzymatic properties. The edited form removes thymine glycol from duplex DNA 30 times more slowly than the form encoded in the genome, whereas editing enhances repair of the guanidinohydantoin lesion by NEIL1. In addition, we show that the NEIL1 recoding site is a preferred editing site for the RNA editing adenosine deaminase ADAR1. The edited adenosine resides in an A-C mismatch in a hairpin stem formed by pairing of exon 6 to the immediate upstream intron 5 sequence. As expected for an ADAR1 site, editing at this position is increased in human cells treated with interferon α. These results suggest a unique regulatory mechanism for DNA repair and extend our understanding of the impact of RNA editing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1.DNA Repair (Amst). 2020 Jan;85:102741. doi: 10.1016/j.dnarep.2019.102741. Epub 2019 Nov 2. DNA Repair (Amst). 2020. PMID: 31733589 Free PMC article.
-
NEIL1 Recoding due to RNA Editing Impacts Lesion-Specific Recognition and Excision.J Am Chem Soc. 2022 Aug 17;144(32):14578-14589. doi: 10.1021/jacs.2c03625. Epub 2022 Aug 2. J Am Chem Soc. 2022. PMID: 35917336 Free PMC article.
-
Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: activity, structure, and the effect of editing.DNA Repair (Amst). 2014 Feb;14:17-26. doi: 10.1016/j.dnarep.2013.12.003. Epub 2013 Dec 29. DNA Repair (Amst). 2014. PMID: 24382305 Free PMC article.
-
Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion.FEBS Lett. 1999 Jun 4;452(1-2):71-6. doi: 10.1016/s0014-5793(99)00590-6. FEBS Lett. 1999. PMID: 10376681 Review.
-
Oxidative DNA damage repair in mammalian cells: a new perspective.DNA Repair (Amst). 2007 Apr 1;6(4):470-80. doi: 10.1016/j.dnarep.2006.10.011. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17116430 Free PMC article. Review.
Cited by
-
A-to-I RNA editing - immune protector and transcriptome diversifier.Nat Rev Genet. 2018 Aug;19(8):473-490. doi: 10.1038/s41576-018-0006-1. Nat Rev Genet. 2018. PMID: 29692414 Review.
-
RNA-Seq analysis identifies a novel set of editing substrates for human ADAR2 present in Saccharomyces cerevisiae.Biochemistry. 2013 Nov 12;52(45):7857-69. doi: 10.1021/bi4006539. Epub 2013 Oct 31. Biochemistry. 2013. PMID: 24124932 Free PMC article.
-
RNA editing by ADAR1 regulates innate and antiviral immune functions in primary macrophages.Sci Rep. 2017 Oct 17;7(1):13339. doi: 10.1038/s41598-017-13580-0. Sci Rep. 2017. PMID: 29042669 Free PMC article.
-
How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs.Bioessays. 2017 Apr;39(4):10.1002/bies.201600187. doi: 10.1002/bies.201600187. Epub 2017 Feb 20. Bioessays. 2017. PMID: 28217931 Free PMC article. Review.
-
Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1.DNA Repair (Amst). 2020 Jan;85:102741. doi: 10.1016/j.dnarep.2019.102741. Epub 2019 Nov 2. DNA Repair (Amst). 2020. PMID: 31733589 Free PMC article.
References
-
- Grosjean H, Benne R. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998.
-
- Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. - PubMed
-
- Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387(6630):303–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

