The twenty-nine amino acid C-terminal cytoplasmic domain of poliovirus 3AB is critical for nucleic acid chaperone activity
- PMID: 21045553
- PMCID: PMC3072266
- DOI: 10.4161/rna.7.6.13781
The twenty-nine amino acid C-terminal cytoplasmic domain of poliovirus 3AB is critical for nucleic acid chaperone activity
Abstract
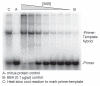
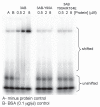
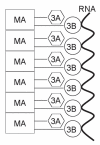
Poliovirus 3AB protein is the first picornavirus protein demonstrated to have nucleic acid chaperone activity. Further characterization of 3AB demonstrates that the C-terminal 22 amino acids (3B region (also referred to as VPg), amino acid 88-109) of the protein is required for chaperone activity, as mutations in this region abrogate nucleic acid binding and chaperone function. Protein 3B alone has no chaperone activity as determined by established assays that include the ability to stimulate nucleic acid hybridization in a primer-template annealing assay, helix-destabilization in a nucleic acid unwinding assay, or aggregation of nucleic acids. In contrast, the putative 3AB C-terminal cytoplasmic domain (C terminal amino acids 81-109, 3B + the last 7 C-terminal amino acids of 3A, termed 3B+7 in this report) possesses strong activity in these assays, albeit at much higher concentrations than 3AB. The characteristics of several mutations in 3B+7 are described here, as well as a model proposing that 3B+7 is the site of the "intrinsic" chaperone activity of 3AB while the 3A N-terminal region (amino acids 1-58) and/or membrane anchor domain (amino acids 59-80) serve to increase the effective concentration of the 3B+7 region leading to the potent chaperone activity of 3AB.
Figures



Similar articles
-
The identification and characterization of nucleic acid chaperone activity of human enterovirus 71 nonstructural protein 3AB.Virology. 2014 Sep;464-465:353-364. doi: 10.1016/j.virol.2014.07.037. Epub 2014 Aug 9. Virology. 2014. PMID: 25113906 Free PMC article.
-
Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase.J Virol. 2007 Jun;81(12):6369-78. doi: 10.1128/JVI.02252-06. Epub 2007 Apr 4. J Virol. 2007. PMID: 17409142 Free PMC article.
-
Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities.J Virol. 2006 Feb;80(4):1662-71. doi: 10.1128/JVI.80.4.1662-1671.2006. J Virol. 2006. PMID: 16439523 Free PMC article.
-
Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism.Prog Nucleic Acid Res Mol Biol. 2005;80:217-86. doi: 10.1016/S0079-6603(05)80006-6. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164976 Review. No abstract available.
-
Roles of intrinsic disorder in protein-nucleic acid interactions.Mol Biosyst. 2012 Jan;8(1):97-104. doi: 10.1039/c1mb05258f. Epub 2011 Aug 26. Mol Biosyst. 2012. PMID: 21874205 Free PMC article. Review.
Cited by
-
The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity.J Virol. 2013 May;87(9):5205-18. doi: 10.1128/JVI.00245-13. Epub 2013 Feb 28. J Virol. 2013. PMID: 23449794 Free PMC article.
-
The identification and characterization of nucleic acid chaperone activity of human enterovirus 71 nonstructural protein 3AB.Virology. 2014 Sep;464-465:353-364. doi: 10.1016/j.virol.2014.07.037. Epub 2014 Aug 9. Virology. 2014. PMID: 25113906 Free PMC article.
-
Norovirus VPg Binds RNA through a Conserved N-Terminal K/R Basic Patch.Viruses. 2021 Jun 30;13(7):1282. doi: 10.3390/v13071282. Viruses. 2021. PMID: 34209211 Free PMC article.
-
Human Enterovirus Nonstructural Protein 2CATPase Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone.PLoS Pathog. 2015 Jul 28;11(7):e1005067. doi: 10.1371/journal.ppat.1005067. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26218680 Free PMC article.
-
The genome-linked protein VPg of vertebrate viruses - a multifaceted protein.Curr Opin Virol. 2011 Nov;1(5):355-62. doi: 10.1016/j.coviro.2011.09.003. Epub 2011 Oct 7. Curr Opin Virol. 2011. PMID: 22440837 Free PMC article. Review.
References
-
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. - PubMed
-
- Levin JG, Guo J, Ioulia R, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acids Res Mol Biol. 2005;80:217–286. - PubMed
-
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
