Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor
- PMID: 21036921
- PMCID: PMC2989995
- DOI: 10.1101/gr.109165.110
Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor
Abstract
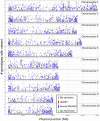
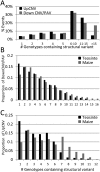
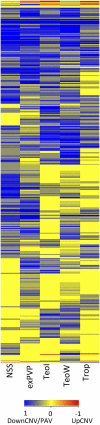
Individuals of the same species are generally thought to have very similar genomes. However, there is growing evidence that structural variation in the form of copy number variation (CNV) and presence-absence variation (PAV) can lead to variation in the genome content of individuals within a species. Array comparative genomic hybridization (CGH) was used to compare gene content and copy number variation among 19 diverse maize inbreds and 14 genotypes of the wild ancestor of maize, teosinte. We identified 479 genes exhibiting higher copy number in some genotypes (UpCNV) and 3410 genes that have either fewer copies or are missing in the genome of at least one genotype relative to B73 (DownCNV/PAV). Many of these DownCNV/PAV are examples of genes present in B73, but missing from other genotypes. Over 70% of the CNV/PAV examples are identified in multiple genotypes, and the majority of events are observed in both maize and teosinte, suggesting that these variants predate domestication and that there is not strong selection acting against them. Many of the genes affected by CNV/PAV are either maize specific (thus possible annotation artifacts) or members of large gene families, suggesting that the gene loss can be tolerated through buffering by redundant functions encoded elsewhere in the genome. While this structural variation may not result in major qualitative variation due to genetic buffering, it may significantly contribute to quantitative variation.
Figures


Similar articles
-
Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content.PLoS Genet. 2009 Nov;5(11):e1000734. doi: 10.1371/journal.pgen.1000734. Epub 2009 Nov 20. PLoS Genet. 2009. PMID: 19956538 Free PMC article.
-
Genetic Architecture of Domestication-Related Traits in Maize.Genetics. 2016 Sep;204(1):99-113. doi: 10.1534/genetics.116.191106. Epub 2016 Jul 13. Genetics. 2016. PMID: 27412713 Free PMC article.
-
Sequence analysis of European maize inbred line F2 provides new insights into molecular and chromosomal characteristics of presence/absence variants.BMC Genomics. 2018 Feb 5;19(1):119. doi: 10.1186/s12864-018-4490-7. BMC Genomics. 2018. PMID: 29402214 Free PMC article.
-
Genomic screening for artificial selection during domestication and improvement in maize.Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review.
-
Genetic, evolutionary and plant breeding insights from the domestication of maize.Elife. 2015 Mar 25;4:e05861. doi: 10.7554/eLife.05861. Elife. 2015. PMID: 25807085 Free PMC article. Review.
Cited by
-
Genes and Small RNA Transcripts Exhibit Dosage-Dependent Expression Pattern in Maize Copy-Number Alterations.Genetics. 2016 Jul;203(3):1133-47. doi: 10.1534/genetics.116.188235. Epub 2016 Apr 29. Genetics. 2016. PMID: 27129738 Free PMC article.
-
Natural Allelic Variations in Highly Polyploidy Saccharum Complex.Front Plant Sci. 2016 Jun 8;7:804. doi: 10.3389/fpls.2016.00804. eCollection 2016. Front Plant Sci. 2016. PMID: 27375658 Free PMC article.
-
Targeted Sequencing Reveals Large-Scale Sequence Polymorphism in Maize Candidate Genes for Biomass Production and Composition.PLoS One. 2015 Jul 7;10(7):e0132120. doi: 10.1371/journal.pone.0132120. eCollection 2015. PLoS One. 2015. PMID: 26151830 Free PMC article.
-
EPAD1 Orthologs Play a Conserved Role in Pollen Exine Patterning.Int J Mol Sci. 2024 Aug 16;25(16):8914. doi: 10.3390/ijms25168914. Int J Mol Sci. 2024. PMID: 39201600 Free PMC article.
-
A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes.BMC Genomics. 2015 Mar 16;16(1):190. doi: 10.1186/s12864-015-1397-4. BMC Genomics. 2015. PMID: 25887443 Free PMC article.
References
-
- Beckmann JS, Estivill X, Antonarakis SE 2007. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet 8: 639–646 - PubMed
-
- Belo A, Beatty MK, Hondred D, Fengler KA, Li B, Rafalski A 2010. Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor Appl Genet 120: 355–367 - PubMed
-
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300
-
- Bennetzen J 2005. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15: 621–627 - PubMed
-
- Bennetzen JL 2007. Patterns in grass genome evolution. Curr Opin Plant Biol 10: 176–181 - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
