Systems vaccinology
- PMID: 21029962
- PMCID: PMC3001343
- DOI: 10.1016/j.immuni.2010.10.006
Systems vaccinology
Abstract
Vaccination is one of the greatest triumphs of modern medicine, yet we remain largely ignorant of the mechanisms by which successful vaccines stimulate protective immunity. Two recent advances are beginning to illuminate such mechanisms: realization of the pivotal role of the innate immune system in sensing microbes and stimulating adaptive immunity, and advances in systems biology. Recent studies have used systems biology approaches to obtain a global picture of the immune responses to vaccination in humans. This has enabled the identification of early innate signatures that predict the immunogenicity of vaccines, and identification of potentially novel mechanisms of immune regulation. Here, we review these advances and critically examine the potential opportunities and challenges posed by systems biology in vaccine development.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



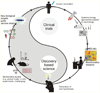
Similar articles
-
Vaccinology in the era of high-throughput biology.Philos Trans R Soc Lond B Biol Sci. 2015 Jun 19;370(1671):20140146. doi: 10.1098/rstb.2014.0146. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25964458 Free PMC article. Review.
-
Systems vaccinology: Enabling rational vaccine design with systems biological approaches.Vaccine. 2015 Sep 29;33(40):5294-301. doi: 10.1016/j.vaccine.2015.03.072. Epub 2015 Apr 6. Vaccine. 2015. PMID: 25858860 Free PMC article. Review.
-
Systems integration of innate and adaptive immunity.Vaccine. 2015 Sep 29;33(40):5241-8. doi: 10.1016/j.vaccine.2015.05.098. Epub 2015 Jun 21. Vaccine. 2015. PMID: 26102534 Review.
-
Systems vaccinology: probing humanity's diverse immune systems with vaccines.Proc Natl Acad Sci U S A. 2014 Aug 26;111(34):12300-6. doi: 10.1073/pnas.1400476111. Epub 2014 Aug 18. Proc Natl Acad Sci U S A. 2014. PMID: 25136102 Free PMC article. Review.
-
Immunological mechanisms of vaccination.Nat Immunol. 2011 Jun;12(6):509-17. doi: 10.1038/ni.2039. Nat Immunol. 2011. PMID: 21739679 Free PMC article. Review.
Cited by
-
A Francisella tularensis live vaccine strain that improves stimulation of antigen-presenting cells does not enhance vaccine efficacy.PLoS One. 2012;7(2):e31172. doi: 10.1371/journal.pone.0031172. Epub 2012 Feb 15. PLoS One. 2012. PMID: 22355343 Free PMC article.
-
Spotlight on systems vaccinology: a novel approach to elucidate correlates of protection.Genes Immun. 2024 Aug;25(4):336-337. doi: 10.1038/s41435-023-00247-2. Epub 2023 Dec 26. Genes Immun. 2024. PMID: 38148341 Free PMC article. No abstract available.
-
Understanding the immune response to seasonal influenza vaccination in older adults: a systems biology approach.Expert Rev Vaccines. 2012 Aug;11(8):985-94. doi: 10.1586/erv.12.61. Expert Rev Vaccines. 2012. PMID: 23002979 Free PMC article. Review.
-
Systems biological approaches to measure and understand vaccine immunity in humans.Semin Immunol. 2013 Oct 31;25(3):209-18. doi: 10.1016/j.smim.2013.05.003. Epub 2013 Jun 21. Semin Immunol. 2013. PMID: 23796714 Free PMC article. Review.
-
Cancer immunotherapy via dendritic cells.Nat Rev Cancer. 2012 Mar 22;12(4):265-77. doi: 10.1038/nrc3258. Nat Rev Cancer. 2012. PMID: 22437871 Free PMC article. Review.
References
-
- Aderem A, Hood L. Immunology in the post-genomic era. Nat Immunol. 2001;2:373–375. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Andreoni J, Kayhty H, Densen P. Vaccination and the role of capsular polysaccharide antibody in prevention of recurrent meningococcal disease in late complement component-deficient individuals. J Infect Dis. 1993;168:227–231. - PubMed
-
- Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

