A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program
- PMID: 20980664
- PMCID: PMC2993380
- DOI: 10.1073/pnas.1007698107
A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program
Abstract
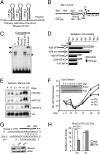
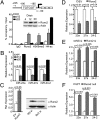
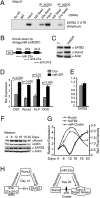
Induced osteogenesis includes a program of microRNAs (miRs) to repress the translation of genes that act as inhibitors of bone formation. How expression of bone-related miRs is regulated remains a compelling question. Here we report that Runx2, a transcription factor essential for osteoblastogenesis, negatively regulates expression of the miR cluster 23a∼27a∼24-2. Overexpression, reporter, and chromatin immunoprecipitation assays established the presence of a functional Runx binding element that represses expression of these miRs. Consistent with this finding, exogenous expression of each of the miRs suppressed osteoblast differentiation, whereas antagomirs increased bone marker expression. The biological significance of Runx2 repression of this miR cluster is that each miR directly targets the 3' UTR of SATB2, which is known to synergize with Runx2 to facilitate bone formation. The findings suggest Runx2-negative regulation of multiple miRs by a feed-forward mechanism to cause derepression of SATB2 to promote differentiation. We find also that miR-23a represses Runx2 in the terminally differentiated osteocyte, representing a feedback mechanism to attenuate osteoblast maturation. We provide direct evidence for an interdependent relationship among transcriptional inhibition of the miR cluster by Runx2, translational repression of Runx2 and of SATB2 by the cluster miRs during progression of osteoblast differentiation. Furthermore, miR cluster gain of function (i.e., inhibition of osteogenesis) is rescued by the exogenous expression of SATB2. Taken together, we have established a regulatory network with a central role for the miR cluster 23a∼27a∼24-2 in both progression and maintenance of the osteocyte phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The microRNA-23a cluster regulates the developmental HoxA cluster function during osteoblast differentiation.J Biol Chem. 2018 Nov 9;293(45):17646-17660. doi: 10.1074/jbc.RA118.003052. Epub 2018 Sep 21. J Biol Chem. 2018. PMID: 30242124 Free PMC article.
-
Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells.Stem Cells Dev. 2013 Aug 15;22(16):2278-86. doi: 10.1089/scd.2012.0686. Epub 2013 Apr 27. Stem Cells Dev. 2013. PMID: 23517179
-
Decrease of MiR-31 induced by TNF-α inhibitor activates SATB2/RUNX2 pathway and promotes osteogenic differentiation in ethanol-induced osteonecrosis.J Cell Physiol. 2019 Apr;234(4):4314-4326. doi: 10.1002/jcp.27210. Epub 2018 Aug 21. J Cell Physiol. 2019. PMID: 30132874
-
Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors.Crit Rev Eukaryot Gene Expr. 2004;14(1-2):1-41. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15104525 Review.
-
Networks and hubs for the transcriptional control of osteoblastogenesis.Rev Endocr Metab Disord. 2006 Jun;7(1-2):1-16. doi: 10.1007/s11154-006-9001-5. Rev Endocr Metab Disord. 2006. PMID: 17051438 Review.
Cited by
-
Fecal Microbiota Transplantation for Recurrent Clostridioides difficile Infection Associates With Functional Alterations in Circulating microRNAs.Gastroenterology. 2021 Jul;161(1):255-270.e4. doi: 10.1053/j.gastro.2021.03.050. Epub 2021 Apr 9. Gastroenterology. 2021. PMID: 33844988 Free PMC article.
-
Antagonizing miR-218-5p attenuates Wnt signaling and reduces metastatic bone disease of triple negative breast cancer cells.Oncotarget. 2016 Nov 29;7(48):79032-79046. doi: 10.18632/oncotarget.12593. Oncotarget. 2016. PMID: 27738322 Free PMC article.
-
Expression of Sp7 in Satb2-induced osteogenic differentiation of mouse bone marrow stromal cells is regulated by microRNA-27a.Mol Cell Biochem. 2016 Jun;417(1-2):7-16. doi: 10.1007/s11010-016-2709-y. Epub 2016 May 3. Mol Cell Biochem. 2016. PMID: 27142530
-
Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression.Stem Cells Dev. 2012 Sep 1;21(13):2531-40. doi: 10.1089/scd.2012.0014. Epub 2012 Apr 2. Stem Cells Dev. 2012. PMID: 22375943 Free PMC article.
-
Vitamin D activation of functionally distinct regulatory miRNAs in primary human osteoblasts.J Bone Miner Res. 2013 Jun;28(6):1478-88. doi: 10.1002/jbmr.1882. J Bone Miner Res. 2013. PMID: 23362149 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

