Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments
- PMID: 20974735
- PMCID: PMC3003824
- DOI: 10.1093/jxb/erq324
Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments
Abstract
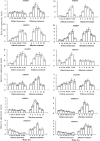
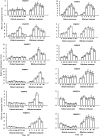
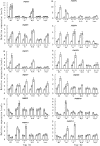
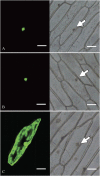
Ethylene-responsive element-binding factor (ERF) genes constitute one of the largest transcription factor gene families in plants. In Arabidopsis and rice, only a few ERF genes have been characterized so far. Flower senescence is associated with increased ethylene production in many flowers. However, the characterization of ERF genes in flower senescence has not been reported. In this study, 13 ERF cDNAs were cloned from petunia. Based on the sequence characterization, these PhERFs could be classified into four of the 12 known ERF families. Their predicted amino acid sequences exhibited similarities to ERFs from other plant species. Expression analyses of PhERF mRNAs were performed in corollas and gynoecia of petunia flower. The 13 PhERF genes displayed differential expression patterns and levels during natural flower senescence. Exogenous ethylene accelerates the transcription of the various PhERF genes, and silver thiosulphate (STS) decreased the transcription of several PhERF genes in corollas and gynoecia. PhERF genes of group VII showed a strong association with the rise in ethylene production in both petals and gynoecia, and might be associated particularly with flower senescence in petunia. The effect of sugar, methyl jasmonate, and the plant hormones abscisic acid, salicylic acid, and 6-benzyladenine in regulating the different PhERF transcripts was investigated. Functional nuclear localization signal analyses of two PhERF proteins (PhERF2 and PhERF3) were carried out using fluorescence microscopy. These results supported a role for petunia PhERF genes in transcriptional regulation of petunia flower senescence processes.
Figures


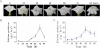

Similar articles
-
A Petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence.PLoS One. 2014 Feb 14;9(2):e88320. doi: 10.1371/journal.pone.0088320. eCollection 2014. PLoS One. 2014. PMID: 24551088 Free PMC article.
-
Functional characterization of PhGR and PhGRL1 during flower senescence in the petunia.Plant Cell Rep. 2015 Sep;34(9):1561-8. doi: 10.1007/s00299-015-1808-7. Epub 2015 May 19. Plant Cell Rep. 2015. PMID: 25987314
-
Overproduction of cytokinins in petunia flowers transformed with P(SAG12)-IPT delays corolla senescence and decreases sensitivity to ethylene.Plant Physiol. 2003 Aug;132(4):2174-83. doi: 10.1104/pp.103.023945. Plant Physiol. 2003. PMID: 12913172 Free PMC article.
-
Regulation of ethylene-induced transcription of defense genes.Plant Cell Physiol. 2000 Nov;41(11):1187-92. doi: 10.1093/pcp/pcd057. Plant Cell Physiol. 2000. PMID: 11092902 Review.
-
Regulation of volatile benzenoid biosynthesis in petunia flowers.Trends Plant Sci. 2006 Jan;11(1):20-5. doi: 10.1016/j.tplants.2005.09.009. Epub 2005 Oct 12. Trends Plant Sci. 2006. PMID: 16226052 Review.
Cited by
-
Mitochondrial Dysfunction, Oxidative Stress and Neuroinflammation in Neurodegeneration with Brain Iron Accumulation (NBIA).Antioxidants (Basel). 2020 Oct 20;9(10):1020. doi: 10.3390/antiox9101020. Antioxidants (Basel). 2020. PMID: 33092153 Free PMC article. Review.
-
PhCESA3 silencing inhibits elongation and stimulates radial expansion in petunia.Sci Rep. 2017 Feb 2;7:41471. doi: 10.1038/srep41471. Sci Rep. 2017. PMID: 28150693 Free PMC article.
-
From models to ornamentals: how is flower senescence regulated?Plant Mol Biol. 2013 Aug;82(6):563-74. doi: 10.1007/s11103-012-9968-0. Epub 2012 Sep 15. Plant Mol Biol. 2013. PMID: 22983713 Review.
-
A Petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence.PLoS One. 2014 Feb 14;9(2):e88320. doi: 10.1371/journal.pone.0088320. eCollection 2014. PLoS One. 2014. PMID: 24551088 Free PMC article.
-
Auxin controls circadian flower opening and closure in the waterlily.BMC Plant Biol. 2018 Jul 11;18(1):143. doi: 10.1186/s12870-018-1357-7. BMC Plant Biol. 2018. PMID: 29996787 Free PMC article.
References
-
- Abeles FB, Morgan PW, Salveit MEJ. Ethylene in plant biology. 2nd edn. San Diego, CA: Academic Press; 1992.
-
- Borochov A, Woodson WR. Physiology and biochemistry of flower petal senescence. Horticultural Reviews. 1989;11:15–43.
-
- Chang YS, Chen HC. Variability between silver thiosulfate and 1-naphthaleneacetic acid applications in prolonging bract longevity of potted bougainvillea. Scientia Horticulturae. 2001;87:217–224.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

