Differential requirement of histone acetylase and deacetylase activities for IRF5-mediated proinflammatory cytokine expression
- PMID: 20935208
- PMCID: PMC3233222
- DOI: 10.4049/jimmunol.1000482
Differential requirement of histone acetylase and deacetylase activities for IRF5-mediated proinflammatory cytokine expression
Abstract
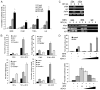
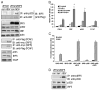
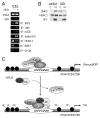
Recent evidence indicates a new role for histone deacetylases (HDACs) in the activation of genes governing the host immune response. Virus, along with other pathogenic stimuli, triggers an antiviral defense mechanism through the induction of IFN, IFN-stimulated genes, and other proinflammatory cytokines. Many of these genes have been shown to be regulated by transcription factors of the IFN regulatory factor (IRF) family. Recent studies from IRF5 knockout mice have confirmed a critical role for IRF5 in virus-induced type I IFN expression and proinflammatory cytokines IL-6, IL-12, and TNF-α; yet, little is known of the molecular mechanism of IRF5-mediated proinflammatory cytokine expression. In this study, we show that both HDACs and histone acetyltransferases (HATs) associate with IRF5, leading to alterations in its transactivation ability. Using the HDAC inhibitor trichostatin A, we demonstrate that ISRE, IFNA, and IL6 promoters require HDAC activity for transactivation and transcription, whereas TNFα does not. Mapping the interaction of corepressor proteins (HDAC1, silencing mediator of retinoid and thyroid receptor/nuclear corepressor of retinoid receptor, and Sin3a) and HATs to IRF5 revealed distinct differences, including the dependence of IRF5 phosphorylation on HAT association resulting in IRF5 acetylation. Data presented in this study support a mechanism whereby virus triggers the dynamic conversion of an IRF5-mediated silencing complex to that of an activating complex on promoters of target genes. These data provide the first evidence, to our knowledge, of a tightly controlled transcriptional mechanism whereby IRF5 regulates proinflammatory cytokine expression in conjunction with HATs and HDACs.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures
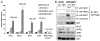


Similar articles
-
IRF5 is elevated in childhood-onset SLE and regulated by histone acetyltransferase and histone deacetylase inhibitors.Oncotarget. 2017 Jul 18;8(29):47184-47194. doi: 10.18632/oncotarget.17586. Oncotarget. 2017. PMID: 28525378 Free PMC article.
-
Epigenetic regulation of high glucose-induced proinflammatory cytokine production in monocytes by curcumin.J Nutr Biochem. 2011 May;22(5):450-8. doi: 10.1016/j.jnutbio.2010.03.014. Epub 2010 Jul 22. J Nutr Biochem. 2011. PMID: 20655188 Free PMC article.
-
Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes.Cell. 2009 Sep 4;138(5):1019-31. doi: 10.1016/j.cell.2009.06.049. Epub 2009 Aug 20. Cell. 2009. PMID: 19698979 Free PMC article.
-
The HAT/HDAC interplay: multilevel control of STAT signaling.Cytokine Growth Factor Rev. 2012 Dec;23(6):283-91. doi: 10.1016/j.cytogfr.2012.08.002. Epub 2012 Sep 16. Cytokine Growth Factor Rev. 2012. PMID: 22989617 Review.
-
Role of histone acetyltransferases and histone deacetylases in adipocyte differentiation and adipogenesis.Eur J Cell Biol. 2014 Apr;93(4):170-7. doi: 10.1016/j.ejcb.2014.03.001. Epub 2014 Mar 25. Eur J Cell Biol. 2014. PMID: 24810880 Review.
Cited by
-
Photochemical control of DNA decoy function enables precise regulation of nuclear factor κB activity.J Am Chem Soc. 2011 Aug 24;133(33):13176-82. doi: 10.1021/ja204980v. Epub 2011 Jul 29. J Am Chem Soc. 2011. PMID: 21761875 Free PMC article.
-
IRF5:RelA interaction targets inflammatory genes in macrophages.Cell Rep. 2014 Sep 11;8(5):1308-17. doi: 10.1016/j.celrep.2014.07.034. Epub 2014 Aug 21. Cell Rep. 2014. PMID: 25159141 Free PMC article.
-
Epigenetic Control of Macrophage Polarisation and Soluble Mediator Gene Expression during Inflammation.Mediators Inflamm. 2016;2016:6591703. doi: 10.1155/2016/6591703. Epub 2016 Apr 10. Mediators Inflamm. 2016. PMID: 27143818 Free PMC article. Review.
-
The COP9 signalosome interacts with and regulates interferon regulatory factor 5 protein stability.Mol Cell Biol. 2013 Mar;33(6):1124-38. doi: 10.1128/MCB.00802-12. Epub 2012 Dec 28. Mol Cell Biol. 2013. Retraction in: Mol Cell Biol. 2017 Nov 28;37(24):e00477-17. doi: 10.1128/MCB.00477-17. PMID: 23275442 Free PMC article. Retracted.
-
Therapeutic Targeting of IRFs: Pathway-Dependence or Structure-Based?Front Immunol. 2018 Nov 20;9:2622. doi: 10.3389/fimmu.2018.02622. eCollection 2018. Front Immunol. 2018. PMID: 30515152 Free PMC article. Review.
References
-
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. - PubMed
-
- Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. - PubMed
-
- Nusinzon I, Horvath CM. Histone deaceylases as transcriptional activators? Role reversal in inducible gene regulation. Sci STKE. 2005;2005:re11. - PubMed
-
- Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. - PubMed
-
- Chambers AE, Banerjee S, Chaplin T, Dunne J, Debernardi S, Joel SP, Young BD. Histone acetylation-mediated regulation of genes in leukaemic cells. Eur J Cancer. 2003;39:1165–1175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

