The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family
- PMID: 20934997
- PMCID: PMC3012774
- DOI: 10.3324/haematol.2010.030924
The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family
Abstract
Background: Polymorphic differences between human leukocyte antigen (HLA) molecules affect the specificity and conformation of their bound peptides and lead to differential selection of the T-cell repertoire. Mismatching during allogeneic transplantation can, therefore, lead to immunological reactions.
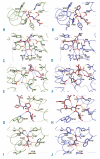
Design and methods: We investigated the structure-function relationships of six members of the HLA-B*41 allelic group that differ by six polymorphic amino acids, including positions 80, 95, 97 and 114 within the antigen-binding cleft. Peptide-binding motifs for B*41:01, *41:02, *41:03, *41:04, *41:05 and *41:06 were determined by sequencing self-peptides from recombinant B*41 molecules by electrospray ionization tandem mass spectrometry. The crystal structures of HLA-B*41:03 bound to a natural 16-mer self-ligand (AEMYGSVTEHPSPSPL) and HLA-B*41:04 bound to a natural 11-mer self-ligand (HEEAVSVDRVL) were solved.
Results: Peptide analysis revealed that all B*41 alleles have an identical anchor motif at peptide position 2 (glutamic acid), but differ in their choice of C-terminal pΩ anchor (proline, valine, leucine). Additionally, B*41:04 displayed a greater preference for long peptides (>10 residues) when compared to the other B*41 allomorphs, while the longest peptide to be eluted from the allelic group (a 16mer) was obtained from B*41:03. The crystal structures of HLA-B*41:03 and HLA-B*41:04 revealed that both alleles interact in a highly conserved manner with the terminal regions of their respective ligands, while micropolymorphism-induced changes in the steric and electrostatic properties of the antigen-binding cleft account for differences in peptide repertoire and auxiliary anchoring.
Conclusions: Differences in peptide repertoire, and peptide length specificity reflect the significant functional evolution of these closely related allotypes and signal their importance in allogeneic transplantation, especially B*41:03 and B*41:04, which accommodate longer peptides, creating structurally distinct peptide-HLA complexes.
Figures

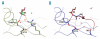
Similar articles
-
Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.J Exp Med. 2009 Jan 16;206(1):209-19. doi: 10.1084/jem.20082136. Epub 2009 Jan 12. J Exp Med. 2009. PMID: 19139173 Free PMC article.
-
Overlap in the repertoires of peptides bound in vivo by a group of related class I HLA-B allotypes.Curr Biol. 1995 Feb 1;5(2):179-90. doi: 10.1016/s0960-9822(95)00039-x. Curr Biol. 1995. PMID: 7743181
-
Identification and characterisation of peptide binding motifs of six autoimmune disease-associated human leukocyte antigen-class I molecules including HLA-B*39:06.Tissue Antigens. 2014 Oct;84(4):378-88. doi: 10.1111/tan.12413. Epub 2014 Aug 25. Tissue Antigens. 2014. PMID: 25154780
-
The impact of HLA-B micropolymorphism outside primary peptide anchor pockets on the CTL response to CMV.Eur J Immunol. 2007 Apr;37(4):946-53. doi: 10.1002/eji.200636588. Eur J Immunol. 2007. PMID: 17357107
-
Function and polymorphism of human leukocyte antigen-A,B,C molecules.Am J Med. 1988 Dec 23;85(6A):2-5. doi: 10.1016/0002-9343(88)90369-5. Am J Med. 1988. PMID: 2462346 Review.
Cited by
-
HLA-E: Presentation of a Broader Peptide Repertoire Impacts the Cellular Immune Response-Implications on HSCT Outcome.Stem Cells Int. 2015;2015:346714. doi: 10.1155/2015/346714. Epub 2015 Aug 12. Stem Cells Int. 2015. PMID: 26366178 Free PMC article.
-
Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation.Front Immunol. 2017 Mar 17;8:292. doi: 10.3389/fimmu.2017.00292. eCollection 2017. Front Immunol. 2017. PMID: 28367149 Free PMC article. Review.
-
Releasing the concept of HLA-allele specific peptide anchors in viral infections: A non-canonical naturally presented human cytomegalovirus-derived HLA-A*24:02 restricted peptide drives exquisite immunogenicity.HLA. 2019 Jul;94(1):25-38. doi: 10.1111/tan.13537. Epub 2019 Apr 14. HLA. 2019. PMID: 30912293 Free PMC article.
-
HLA-B*57 Micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control.J Virol. 2012 Jan;86(2):919-29. doi: 10.1128/JVI.06150-11. Epub 2011 Nov 16. J Virol. 2012. PMID: 22090105 Free PMC article.
-
Naturally processed non-canonical HLA-A*02:01 presented peptides.J Biol Chem. 2015 Jan 30;290(5):2593-603. doi: 10.1074/jbc.M114.607028. Epub 2014 Dec 12. J Biol Chem. 2015. PMID: 25505266 Free PMC article.
References
-
- Chelvanayagam G. A roadmap for HLA-A, HLA-B, and HLA-C peptide binding specificities. Immunogenetics. 1996;45(1):15–26. - PubMed
-
- Zhao Y, Gran B, Pinilla C, Markovic-Plese S, Hemmer B, Tzou A, et al. Combinatorial peptide libraries and biometric score matrices permit the quantitative analysis of specific and degenerate interactions between clonotypic TCR and MHC peptide ligands. J Immunol. 2001;167(4):2130–41. - PubMed
-
- Zhang C, Bickis MG, Wu FX, Kusalik AJ. Optimally-connected hidden Markov models for predicting MHC-binding peptides. J Bioinform Comput Biol. 2006;4(5):959–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

