Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster
- PMID: 20929442
- PMCID: PMC2992795
- DOI: 10.1042/BJ20101440
Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster
Abstract
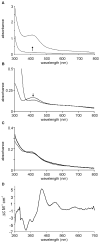
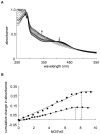
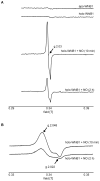
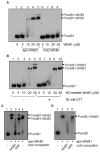
Mycobacterium tuberculosis is a major pathogen that has the ability to establish, and emerge from, a persistent state. Wbl family proteins are associated with developmental processes in actinomycetes, and M. tuberculosis has seven such proteins. In the present study it is shown that the M. tuberculosis H37Rv whiB1 gene is essential. The WhiB1 protein possesses a [4Fe-4S]2+ cluster that is stable in air but reacts rapidly with eight equivalents of nitric oxide to yield two dinuclear dinitrosyl-iron thiol complexes. The [4Fe-4S] form of WhiB1 did not bind whiB1 promoter DNA, but the reduced and oxidized apo-WhiB1, and nitric oxide-treated holo-WhiB1 did bind to DNA. Mycobacterium smegmatis RNA polymerase induced transcription of whiB1 in vitro; however, in the presence of apo-WhiB1, transcription was severely inhibited, irrespective of the presence or absence of the CRP (cAMP receptor protein) Rv3676, which is known to activate whiB1 expression. Footprinting suggested that autorepression of whiB1 is achieved by apo-WhiB1 binding at a region that overlaps the core promoter elements. A model incorporating regulation of whiB1 expression in response to nitric oxide and cAMP is discussed with implications for sensing two important signals in establishing M. tuberculosis infections.
Figures





Similar articles
-
Structure-function relationships of the Mycobacterium tuberculosis transcription factor WhiB1.PLoS One. 2012;7(7):e40407. doi: 10.1371/journal.pone.0040407. Epub 2012 Jul 5. PLoS One. 2012. PMID: 22792304 Free PMC article.
-
Mycobacterium tuberculosis WhiB1 represses transcription of the essential chaperonin GroEL2.Tuberculosis (Edinb). 2012 Jul;92(4):328-32. doi: 10.1016/j.tube.2012.03.001. Epub 2012 Mar 29. Tuberculosis (Edinb). 2012. PMID: 22464736 Free PMC article.
-
Regulation of the expression of whiB1 in Mycobacterium tuberculosis: role of cAMP receptor protein.Microbiology (Reading). 2006 Sep;152(Pt 9):2749-2756. doi: 10.1099/mic.0.28924-0. Microbiology (Reading). 2006. PMID: 16946269
-
Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide.Acc Chem Res. 2014 Oct 21;47(10):3196-205. doi: 10.1021/ar5002507. Epub 2014 Sep 29. Acc Chem Res. 2014. PMID: 25262769 Review.
-
Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli.Methods Enzymol. 2008;437:191-209. doi: 10.1016/S0076-6879(07)37011-0. Methods Enzymol. 2008. PMID: 18433630 Review.
Cited by
-
Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis.PLoS Pathog. 2014 Mar 6;10(3):e1003994. doi: 10.1371/journal.ppat.1003994. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24603869 Free PMC article.
-
Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches.Curr Opin Microbiol. 2014 Apr;18(100):1-7. doi: 10.1016/j.mib.2014.01.003. Epub 2014 Feb 7. Curr Opin Microbiol. 2014. PMID: 24509484 Free PMC article. Review.
-
WhiB-like proteins: Diversity of structure, function and mechanism.Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119787. doi: 10.1016/j.bbamcr.2024.119787. Epub 2024 Jun 13. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38879133 Review.
-
Convergence of two global regulators to coordinate expression of essential virulence determinants of Mycobacterium tuberculosis.Elife. 2022 Nov 9;11:e80965. doi: 10.7554/eLife.80965. Elife. 2022. PMID: 36350294 Free PMC article.
-
An intricate regulation of WblA controlling production of silent tylosin analogues and abolishment of expressible nikkomycin.Sci China Life Sci. 2023 Mar;66(3):612-625. doi: 10.1007/s11427-022-2199-1. Epub 2023 Jan 4. Sci China Life Sci. 2023. PMID: 36607495
References
-
- World Health Organization . Global Tuberculosis Control: a short update to the 2009 report. WHO Press; Geneva Switzerland: 2009. WHO/HTM/TB/2009.426.
-
- Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2001;2:569–577. - PubMed
-
- Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 2003;1:97–105. - PubMed
-
- Rickman L, Scott C, Hunt DM, Hutchinson T, Menendez MC, Whalan R, Hinds J, Colston MJ, Green J, Buxton RS. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 2005;56:1274–1286. - PMC - PubMed
-
- Stapleton MR, Haq I, Hunt DM, Arnvig KB, Artymiuk PJ, Buxton RS, Green J. Mycobacterium tuberculosis cAMP receptor protein (Rv3676) differs from the Escherichia coli paradigm in its cAMP binding and DNA binding properties and transcription activation properties. J. Biol. Chem. 2010;285:7016–7027. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

