Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi
- PMID: 20920349
- PMCID: PMC2959043
- DOI: 10.1186/2041-9139-1-10
Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi
Abstract
Background: Intercellular signaling pathways are a fundamental component of the integrating cellular behavior required for the evolution of multicellularity. The genomes of three of the four early branching animal phyla (Cnidaria, Placozoa and Porifera) have been surveyed for key components, but not the fourth (Ctenophora). Genomic data from ctenophores could be particularly relevant, as ctenophores have been proposed to be one of the earliest branching metazoan phyla.
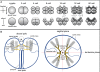
Results: A preliminary assembly of the lobate ctenophore Mnemiopsis leidyi genome generated using next-generation sequencing technologies were searched for components of a developmentally important signaling pathway, the Wnt/β-catenin pathway. Molecular phylogenetic analysis shows four distinct Wnt ligands (MlWnt6, MlWnt9, MlWntA and MlWntX), and most, but not all components of the receptor and intracellular signaling pathway were detected. In situ hybridization of the four Wnt ligands showed that they are expressed in discrete regions associated with the aboral pole, tentacle apparati and apical organ.
Conclusions: Ctenophores show a minimal (but not obviously simple) complement of Wnt signaling components. Furthermore, it is difficult to compare the Mnemiopsis Wnt expression patterns with those of other metazoans. mRNA expression of Wnt pathway components appears later in development than expected, and zygotic gene expression does not appear to play a role in early axis specification. Notably absent in the Mnemiopsis genome are most major secreted antagonists, which suggests that complex regulation of this secreted signaling pathway probably evolved later in animal evolution.
Figures





Similar articles
-
Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi.PLoS One. 2011;6(9):e24152. doi: 10.1371/journal.pone.0024152. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931657 Free PMC article.
-
The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa.Evodevo. 2010 Oct 4;1(1):9. doi: 10.1186/2041-9139-1-9. Evodevo. 2010. PMID: 20920347 Free PMC article.
-
Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily?Evodevo. 2011 Feb 3;2(1):3. doi: 10.1186/2041-9139-2-3. Evodevo. 2011. PMID: 21291545 Free PMC article.
-
Whole-Body Regeneration in the Lobate Ctenophore Mnemiopsis leidyi.Genes (Basel). 2021 Jun 5;12(6):867. doi: 10.3390/genes12060867. Genes (Basel). 2021. PMID: 34198839 Free PMC article. Review.
-
Brief History of Ctenophora.Methods Mol Biol. 2024;2757:1-26. doi: 10.1007/978-1-0716-3642-8_1. Methods Mol Biol. 2024. PMID: 38668961 Review.
Cited by
-
Retracing the path of planar cell polarity.BMC Evol Biol. 2016 Apr 2;16:69. doi: 10.1186/s12862-016-0641-0. BMC Evol Biol. 2016. PMID: 27039172 Free PMC article.
-
Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development.PLoS Biol. 2018 Jan 16;16(1):e2003698. doi: 10.1371/journal.pbio.2003698. eCollection 2018 Jan. PLoS Biol. 2018. PMID: 29337984 Free PMC article.
-
Controversies surrounding segments and parasegments in onychophora: insights from the expression patterns of four "segment polarity genes" in the peripatopsid Euperipatoides rowelli.PLoS One. 2014 Dec 3;9(12):e114383. doi: 10.1371/journal.pone.0114383. eCollection 2014. PLoS One. 2014. PMID: 25470738 Free PMC article.
-
The evolution of the Wnt pathway.Cold Spring Harb Perspect Biol. 2012 Jul 1;4(7):a007922. doi: 10.1101/cshperspect.a007922. Cold Spring Harb Perspect Biol. 2012. PMID: 22751150 Free PMC article.
-
Apcdd1 is a dual BMP/Wnt inhibitor in the developing nervous system and skin.Dev Biol. 2020 Aug 1;464(1):71-87. doi: 10.1016/j.ydbio.2020.03.015. Epub 2020 Apr 19. Dev Biol. 2020. PMID: 32320685 Free PMC article.
References
-
- Nielsen C, Scharff N, Eibye-Jacobsen D. Cladistic analyses of the animal kingdom. Biol J Linn Soc. 1996;57:385–410. doi: 10.1111/j.1095-8312.1996.tb01857.x. - DOI
-
- Zrzavy J, Mihulka S, Kepka P, Bezdek A, Tietz D. Phylogeny of the Metazoa based on morphological and 18S ribosomal DNA evidence. Cladistics. 1998;14:249–285. - PubMed
-
- Kim J, Kim W, Cunningham CW. A new perspective on lower metazoan relationships from 18S rDNA sequences. Mol Biol Evol. 1999;16:423–427. - PubMed
LinkOut - more resources
Full Text Sources

